Nucleosome translational position, not histone acetylation, determines TFIIIA binding to nucleosomal Xenopus laevis 5S rRNA genes
- PMID: 9488430
- PMCID: PMC108828
- DOI: 10.1128/MCB.18.3.1156
Nucleosome translational position, not histone acetylation, determines TFIIIA binding to nucleosomal Xenopus laevis 5S rRNA genes
Abstract
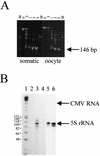
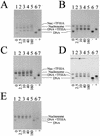
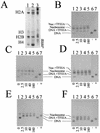
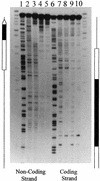
We sought to study the binding constraints placed on the nine-zinc-finger protein transcription factor IIIA (TFIIIA) by a histone octamer. To this end, five overlapping fragments of the Xenopus laevis oocyte and somatic 5S rRNA genes were reconstituted into nucleosomes, and it was subsequently shown that nucleosome translational positioning is a major determinant of the binding of TFIIIA to the 5S rRNA genes. Furthermore, it was found that histone acetylation cannot override the TFIIIA binding constraints imposed by unfavorable translational positions.
Figures


References
-
- Ausió J. Structure and dynamics of transcriptionally active chromatin. J Cell Sci. 1992;102:1–5. - PubMed
-
- Ausió J, Dong F, van Holde K E. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone “tails” in the stabilization of the nucleosome. J Mol Biol. 1989;206:451–463. - PubMed
-
- Ausió J, Seger D S, Eisenberg H. Nucleosome core particle stability and conformational change. J Mol Biol. 1984;176:77–104. - PubMed
-
- Ausió J, van Holde K E. Histone hyperacetylation: its effects on nucleosome conformation and stability. Biochemistry. 1986;25:1421–1428. - PubMed
-
- Blomquist P, Li Q, Wrange O. The affinity of nuclear factor 1 for its DNA site is drastically reduced by nucleosome organization irrespective of its rotational or translational position. J Biol Chem. 1996;271:153–159. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
