Two members of the Tcf family implicated in Wnt/beta-catenin signaling during embryogenesis in the mouse
- PMID: 9488439
- PMCID: PMC108837
- DOI: 10.1128/MCB.18.3.1248
Two members of the Tcf family implicated in Wnt/beta-catenin signaling during embryogenesis in the mouse
Abstract
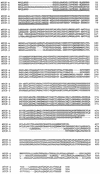
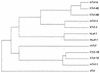
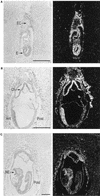
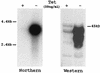
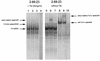
Tcf transcription factors interact with beta-catenin and Armadillo to mediate Wnt/Wingless signaling. We now report the characterization of genes encoding two murine members of the Tcf family, mTcf-3 and mTcf-4. mTcf-3 mRNA is ubiquitously present in embryonic day 6.5 (E6.5) mouse embryos but gradually disappears over the next 3 to 4 days. mTcf-4 expression occurs first at E10.5 and is restricted to di- and mesencephalon and the intestinal epithelium during embryogenesis. The mTcf-3 and mTcf-4 proteins bind a canonical Tcf DNA motif and can complex with the transcriptional coactivator beta-catenin. Overexpression of Wnt-1 in a mammary epithelial cell line leads to the formation of a nuclear complex between beta-catenin and Tcf proteins and to Tcf reporter gene transcription. These data demonstrate a direct link between Wnt stimulation and beta-catenin/Tcf transcriptional activation and imply a role for mTcf-3 and -4 in early Wnt-driven developmental decisions in the mouse embryo.
Figures

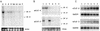

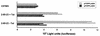
References
-
- Barker, N. Unpublished data.
-
- Behrens J, von Kries J P, Kuehl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of β-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. - PubMed
-
- Brown A M, Wildin R S, Prendergast T J, Varmus H E. A retrovirus vector expressing the putative mammary oncogen int-1 causes partial transformation of a mammary epithelial cell line. Cell. 1986;46:1001–1009. - PubMed
-
- Brunner E, Peter O, Schweizer L, Basler K. Pangolin encodes a Lef-1 homologue that acts downstream of Armadillo to transduce the Wingless signal in Drosophila. Nature. 1997;385:829–833. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
