Regulation of the p85/p110 phosphatidylinositol 3'-kinase: stabilization and inhibition of the p110alpha catalytic subunit by the p85 regulatory subunit
- PMID: 9488453
- PMCID: PMC108851
- DOI: 10.1128/MCB.18.3.1379
Regulation of the p85/p110 phosphatidylinositol 3'-kinase: stabilization and inhibition of the p110alpha catalytic subunit by the p85 regulatory subunit
Abstract
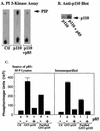
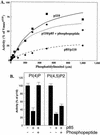
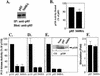
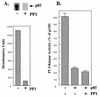
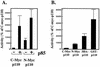
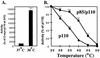
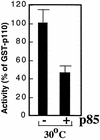
We propose a novel model for the regulation of the p85/pl10alpha phosphatidylinositol 3'-kinase. In insect cells, the p110alpha catalytic subunit is active as a monomer but its activity is decreased by coexpression with the p85 regulatory subunit. Similarly, the lipid kinase activity of recombinant glutathione S-transferase (GST)-p110alpha is reduced by 65 to 85% upon in vitro reconstitution with p85. Incubation of p110alpha/p85 dimers with phosphotyrosyl peptides restored activity, but only to the level of monomeric p110alpha. These data show that the binding of phosphoproteins to the SH2 domains of p85 activates the p85/p110alpha dimers by inducing a transition from an inhibited to a disinhibited state. In contrast, monomeric p110 had little activity in HEK 293T cells, and its activity was increased 15- to 20-fold by coexpression with p85. However, this apparent requirement for p85 was eliminated by the addition of a bulky tag to the N terminus of p110alpha or by the growth of the HEK 293T cells at 30 degrees C. These nonspecific interventions mimicked the effects of p85 on p110alpha, suggesting that the regulatory subunit acts by stabilizing the overall conformation of the catalytic subunit rather than by inducing a specific activated conformation. This stabilization was directly demonstrated in metabolically labeled HEK 293T cells, in which p85 increased the half-life of p110. Furthermore, p85 protected p110 from thermal inactivation in vitro. Importantly, when we examined the effect of p85 on GST-p110alpha in mammalian cells at 30 degrees C, culture conditions that stabilize the catalytic subunit and that are similar to the conditions used for insect cells, we found that p85 inhibited p110alpha. Thus, we have experimentally distinguished two effects of p85 on p110alpha: conformational stabilization of the catalytic subunit and inhibition of its lipid kinase activity. Our data reconcile the apparent conflict between previous studies of insect versus mammalian cells and show that p110alpha is both stabilized and inhibited by dimerization with p85.
Figures


References
-
- Booker G W, Gout I, Downing A K, Driscoll P C, Boyd J, Waterfield M D, Campbell I D. Solution structure and ligand-binding site of the SH3 domain of the p85α subunit of phosphatidylinositol 3-kinase. Cell. 1993;73:813–822. - PubMed
-
- Cantley L C, Auger K R, Carpenter C, Duckworth B, Graziani A, Kapeller R, Soltoff S. Oncogenes and signal transduction. Cell. 1991;64:231–302. - PubMed
-
- Carpenter C L, Auger K R, Chanudhuri M, Yoakim M, Schaffhausen B, Shoelson S, Cantley L C. Phosphoinositide 3-kinase is activated by phosphopeptides that bind to the SH2 domains of the 85-kDa subunit. J Biol Chem. 1993;268:9478–9483. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
