Initiator-elongator discrimination in vertebrate tRNAs for protein synthesis
- PMID: 9488462
- PMCID: PMC108860
- DOI: 10.1128/MCB.18.3.1459
Initiator-elongator discrimination in vertebrate tRNAs for protein synthesis
Abstract
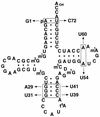
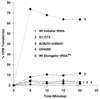
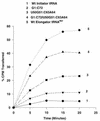
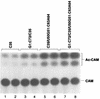
Initiator tRNAs are used exclusively for initiation of protein synthesis and not for the elongation step. We show, in vivo and in vitro, that the primary sequence feature that prevents the human initiator tRNA from acting in the elongation step is the nature of base pairs 50:64 and 51:63 in the TpsiC stem of the initiator tRNA. Various considerations suggest that this is due to sequence-dependent perturbation of the sugar phosphate backbone in the TpsiC stem of initiator tRNA, which most likely blocks binding of the elongation factor to the tRNA. Because the sequences of all vertebrate initiator tRNAs are identical, our findings with the human initiator tRNA are likely to be valid for all vertebrate systems. We have developed reporter systems that can be used to monitor, in mammalian cells, the activity in elongation of mutant human initiator tRNAs carrying anticodon sequence mutations from CAU to CCU (the C35 mutant) or to CUA (the U35A36 mutant). Combination of the anticodon sequence mutation with mutations in base pairs 50:64 and 51:63 yielded tRNAs that act as elongators in mammalian cells. Further mutation of the A1:U72 base pair, which is conserved in virtually all eukaryotic initiator tRNAs, to G1:C72 in the C35 mutant background yielded tRNAs that were even more active in elongation. In addition, in a rabbit reticulocyte in vitro protein-synthesizing system, a tRNA carrying the TpsiC stem and the A1:U72-to-G1:C72 mutations was almost as active in elongation as the elongator methionine tRNA. The combination of mutant initiator tRNA with the CCU anticodon and the reporter system developed here provides the first example of missense suppression in mammalian cells.
Figures


References
-
- Åström S U, Byström A S. Rit1, a tRNA backbone-modifying enzyme that mediates initiator and elongator tRNA discrimination. Cell. 1994;79:535–546. - PubMed
-
- Åström S U, von Pawel-Rammingen U, Byström A S. The yeast initiator tRNAMet can act as an elongator tRNAMetin vivo. J Mol Biol. 1993;233:43–58. - PubMed
-
- Baron C, Böck A. The selenocysteine-inserting tRNA species: structure and function. In: Söll D, RajBhandary U L, editors. Transfer RNA: structure, biosynthesis, and function. Washington, D.C: American Society for Microbiology; 1995. pp. 529–544.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
