4-Coumarate:coenzyme A ligase in hybrid poplar. Properties of native enzymes, cDNA cloning, and analysis of recombinant enzymes
- PMID: 9489021
- PMCID: PMC35134
- DOI: 10.1104/pp.116.2.743
4-Coumarate:coenzyme A ligase in hybrid poplar. Properties of native enzymes, cDNA cloning, and analysis of recombinant enzymes
Abstract
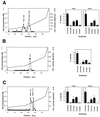
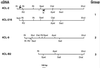
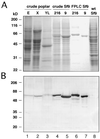
The enzyme 4-coumarate:coenzyme A ligase (4CL) is important in providing activated thioester substrates for phenylpropanoid natural product biosynthesis. We tested different hybrid poplar (Populus trichocarpa x Populus deltoides) tissues for the presence of 4CL isoforms by fast-protein liquid chromatography and detected a minimum of three 4CL isoforms. These isoforms shared similar hydroxycinnamic acid substrate-utilization profiles and were all inactive against sinapic acid, but instability of the native forms precluded extensive further analysis. 4CL cDNA clones were isolated and grouped into two major classes, the predicted amino acid sequences of which were 86% identical. Genomic Southern blots showed that the cDNA classes represent two poplar 4CL genes, and northern blots provided evidence for their differential expression. Recombinant enzymes corresponding to the two genes were expressed using a baculovirus system. The two recombinant proteins had substrate utilization profiles similar to each other and to the native poplar 4CL isoforms (4-coumaric acid > ferulic acid > caffeic acid; there was no conversion of sinapic acid), except that both had relatively high activity toward cinnamic acid. These results are discussed with respect to the role of 4CL in the partitioning of carbon in phenylpropanoid metabolism.
Figures




References
-
- Allina SM, Douglas CJ. Isolation and characterization of the 4-coumarate: CoA ligase gene family in a poplar hybrid (abstract No. 852) Plant Physiol. 1994;105:S-154.
-
- Becker-André M, Schultze-Lefert P, Hahlbrock K. Structural comparison, modes of expression, and putative cis-acting elements of the two 4-coumarate:CoA ligase genes in potato. J Biol Chem. 1991;266:8551–8559. - PubMed
-
- Bradshaw H, Jr, Villar M, Watson B, Otto K, Stewart S, Stettler R. Molecular genetics of growth and development in Populus. III. A genetic linkage map of a hybrid poplar composed of RFLP, STS, and RAPD markers. Theor Appl Genet. 1994;89:167–178. - PubMed
-
- Bradshaw HD, Jr, Stettler RF. Molecular genetics of growth and development in Populus. I. Triploidy in hybrid poplars. Theor Appl Genet. 1993;86:301–307. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

