Interphase cell cycle dynamics of a late-replicating, heterochromatic homogeneously staining region: precise choreography of condensation/decondensation and nuclear positioning
- PMID: 9490713
- PMCID: PMC2132695
- DOI: 10.1083/jcb.140.5.975
Interphase cell cycle dynamics of a late-replicating, heterochromatic homogeneously staining region: precise choreography of condensation/decondensation and nuclear positioning
Abstract
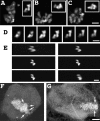
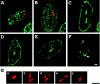
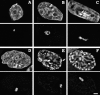
Recently we described a new method for in situ localization of specific DNA sequences, based on lac operator/repressor recognition (Robinett, C.C., A. Straight, G. Li, C. Willhelm, G. Sudlow, A. Murray, and A.S. Belmont. 1996. J. Cell Biol. 135:1685-1700). We have applied this methodology to visualize the cell cycle dynamics of an approximately 90 Mbp, late-replicating, heterochromatic homogeneously staining region (HSR) in CHO cells, combining immunostaining with direct in vivo observations. Between anaphase and early G1, the HSR extends approximately twofold to a linear, approximately 0.3-mum-diam chromatid, and then recondenses to a compact mass adjacent to the nuclear envelope. No further changes in HSR conformation or position are seen through mid-S phase. However, HSR DNA replication is preceded by a decondensation and movement of the HSR into the nuclear interior 4-6 h into S phase. During DNA replication the HSR resolves into linear chromatids and then recondenses into a compact mass; this is followed by a third extension of the HSR during G2/ prophase. Surprisingly, compaction of the HSR is extremely high at all stages of interphase. Preliminary ultrastructural analysis of the HSR suggests at least three levels of large-scale chromatin organization above the 30-nm fiber.
Figures







References
-
- Becker, W.M., J.B. Reece, and M.F. Poenie. 1996. The World of the Cell. The Benjamin/Cummings Publishing Co., Menlo Park. 434–435.
-
- Belmont, A.S. 1997. Large–scale chromatin organization. In Genome Structure and Function. Kluwer Academic Publishers, Dordrecht, Germany. 261–276.
-
- Belmont AS, Braunfeld MB, Sedat JW, Agard DA. Large-scale chromatin structural domains within mitotic and interphase chromosomes in vivo and in vitro. Chromosoma. 1989;98:129–143. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

