Salmonella typhimurium encodes an SdiA homolog, a putative quorum sensor of the LuxR family, that regulates genes on the virulence plasmid
- PMID: 9495757
- PMCID: PMC107006
- DOI: 10.1128/JB.180.5.1185-1193.1998
Salmonella typhimurium encodes an SdiA homolog, a putative quorum sensor of the LuxR family, that regulates genes on the virulence plasmid
Abstract
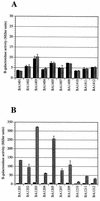
Quorum sensing is a phenomenon in which bacteria sense and respond to their own population density by releasing and sensing pheromones. In gram-negative bacteria, quorum sensing is often performed by the LuxR family of transcriptional regulators, which affect phenotypes as diverse as conjugation, bioluminescence, and virulence gene expression. The gene encoding one LuxR family member, named sdiA (suppressor of cell division inhibition), is present in the Escherichia coli genome. In this report, we have cloned the Salmonella typhimurium homolog of SdiA and performed a systematic screen for sdiA-regulated genes. A 4.4-kb fragment encoding the S. typhimurium sdiA gene was sequenced and found to encode the 3' end of YecC (homologous to amino acid transporters of the ABC family), all of SdiA and SirA (Salmonella invasion regulator), and the 5' end of UvrC. This gene organization is conserved between E. coli and S. typhimurium. We determined that the S. typhimurium sdiA gene was able to weakly complement the E. coli sdiA gene for activation of ftsQAZ at promoter 2 and for suppression of filamentation caused by an ftsZ(Ts) allele. To better understand the function of sdiA in S. typhimurium, we screened 10,000 random lacZY transcriptional fusions (MudJ transposon mutations) for regulation by sdiA. Ten positively regulated fusions were isolated. Seven of the fusions were within an apparent operon containing ORF8, ORF9, rck (resistance to complement killing), and ORF11 of the S. typhimurium virulence plasmid. The three ORFs have now been named srgA, srgB, and srgC (for sdiA-regulated gene), respectively. The DNA sequence adjacent to the remaining three fusions shared no similarity with previously described genes.
Figures




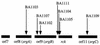
References
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1995.
-
- Bajaj V, Lucas R L, Hwang C, Lee C A. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol. 1996;22:703–714. - PubMed
-
- Bardwell J C, McGovern K, Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991;67:581–589. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

