Insertional inactivation of genes encoding the crystalline inclusion proteins of Photorhabdus luminescens results in mutants with pleiotropic phenotypes
- PMID: 9495767
- PMCID: PMC107016
- DOI: 10.1128/JB.180.5.1261-1269.1998
Insertional inactivation of genes encoding the crystalline inclusion proteins of Photorhabdus luminescens results in mutants with pleiotropic phenotypes
Abstract
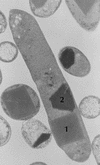
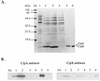
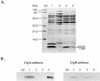
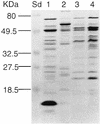
The entomopathogenic bacterium Photorhabdus luminescens exhibits phase variation when cultured in vitro. The variant forms of P. luminescens are pleiotropic and are designated phase I and phase II variants. One of the characteristic phenotypes of phase I cells is the production of two types of intracellular protein inclusions. The genes encoding the protein monomers that form these inclusions, designated cipA and cipB, were cloned and characterized. cipA and cipB encode hydrophobic proteins of 11,648 and 11,308 Da, respectively. The deduced amino acid sequences of CipA and CipB have no significant amino acid sequence similarity to any other known protein but have 25% identity and 49% similarity to each other. Insertional inactivation of cipA or cipB in phase I cells of P. luminescens produced mutants that differ from phase I cells in bioluminescence, the pattern and activities of extracellular products, biochemical traits, adsorption of dyes, and ability to support nematode growth and reproduction. In general, the cip mutants were phenotypically more similar to each other than to either phase I or phase II variants.
Figures



References
-
- Akhurst R J. Morphological and functional phase variation in Xenorhabdus spp.: bacteria symbiotically associated with the insect pathogenic nematodes Neoplectana and Heterorhabditis. J Gen Microbiol. 1980;121:303–309.
-
- Akhurst R J. Antibiotic activity of Xenorhabdus spp.: bacteria symbiotically associated with insect pathogenic nematodes of the families Heterorhabditidae and Steinernematidae. J Gen Microbiol. 1982;128:3061–3065. - PubMed
-
- Akhurst R J, Boemare N E. A numerical taxonomic study of the genus Xenorhabdus (Enterobacteriaceae) and proposed elevation of the subspecies of X. nematophilus to species. J Gen Microbiol. 1988;134:1835–1845. - PubMed
-
- Akhurst, R. J. Personal communication.
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

