Competition for DNA binding sites between the short and long forms of E2 dimers underlies repression in bovine papillomavirus type 1 DNA replication control
- PMID: 9499046
- PMCID: PMC109485
- DOI: 10.1128/JVI.72.3.1931-1940.1998
Competition for DNA binding sites between the short and long forms of E2 dimers underlies repression in bovine papillomavirus type 1 DNA replication control
Abstract
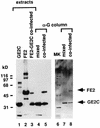
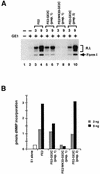
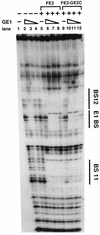
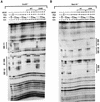
Papillomaviruses establish a long-term latency in vivo by maintaining their genomes as nuclear plasmids in proliferating cells. Bovine papillomavirus type 1 encodes two proteins required for viral DNA replication: the helicase E1 and the positive regulator E2. The homodimeric E2 is known to cooperatively bind to DNA with E1 to form a preinitiation complex at the origin of DNA replication. The virus also codes for two short forms of E2 that can repress viral functions when overexpressed, and at least one copy of the repressor is required for stable plasmid maintenance in transformed cells. Employing a tetracycline-regulated system to control E1 and E2 production from integrated loci, we show that the short form of E2 negatively regulates DNA replication. We also found that the short form could repress replication in a cell-free replication system and that the repression requires the DNA binding domain of the protein. In contrast, heterodimers of the short and long forms were activators and, by footprint analysis, were shown to be as potent as homodimeric E2 in loading E1 to its cognate site. DNA binding studies show that when E1 levels are low and are dependent upon E2 for occupancy of the origin site, the repressor can block E1-DNA interactions. We conclude that DNA replication modulation results from competition between the different forms of E2 for DNA binding. Given that heterodimers are active and that the repressor form of E2 shows little cooperativity with E1 for DNA binding, this protein is a weak repressor.
Figures




Similar articles
-
Characterization of the functional activities of the bovine papillomavirus type 1 E2 protein single-chain heterodimers.J Virol. 2006 Nov;80(22):11218-25. doi: 10.1128/JVI.01127-06. Epub 2006 Aug 30. J Virol. 2006. PMID: 16943289 Free PMC article.
-
Binding of the E1 and E2 proteins to the origin of replication of bovine papillomavirus.J Virol. 1997 Apr;71(4):2887-96. doi: 10.1128/JVI.71.4.2887-2896.1997. J Virol. 1997. PMID: 9060646 Free PMC article.
-
Bovine papillomavirus type 1 E2 protein heterodimer is functional in papillomavirus DNA replication in vivo.Virology. 2009 Apr 10;386(2):353-9. doi: 10.1016/j.virol.2009.01.025. Epub 2009 Feb 20. Virology. 2009. PMID: 19232665
-
Papillomavirus E1 proteins: form, function, and features.Virus Genes. 2002 Jun;24(3):275-90. doi: 10.1023/a:1015336817836. Virus Genes. 2002. PMID: 12086149 Review.
-
The functions of papillomavirus E2 proteins.Virology. 2025 Feb;603:110387. doi: 10.1016/j.virol.2024.110387. Epub 2024 Dec 31. Virology. 2025. PMID: 39826199 Review.
Cited by
-
Characterization of the functional activities of the bovine papillomavirus type 1 E2 protein single-chain heterodimers.J Virol. 2006 Nov;80(22):11218-25. doi: 10.1128/JVI.01127-06. Epub 2006 Aug 30. J Virol. 2006. PMID: 16943289 Free PMC article.
-
The papillomavirus E8-E2C protein represses DNA replication from extrachromosomal origins.Mol Cell Biol. 2003 Nov;23(22):8352-62. doi: 10.1128/MCB.23.22.8352-8362.2003. Mol Cell Biol. 2003. PMID: 14585992 Free PMC article.
-
The papillomavirus E2 proteins.Virology. 2013 Oct;445(1-2):57-79. doi: 10.1016/j.virol.2013.06.006. Epub 2013 Jul 10. Virology. 2013. PMID: 23849793 Free PMC article. Review.
-
Two patches of amino acids on the E2 DNA binding domain define the surface for interaction with E1.J Virol. 2000 Feb;74(3):1506-12. doi: 10.1128/jvi.74.3.1506-1512.2000. J Virol. 2000. PMID: 10627562 Free PMC article.
-
The hinge of the human papillomavirus type 11 E2 protein contains major determinants for nuclear localization and nuclear matrix association.J Virol. 2000 Apr;74(8):3761-70. doi: 10.1128/jvi.74.8.3761-3770.2000. J Virol. 2000. PMID: 10729151 Free PMC article.
References
-
- Cheng S, Schmidt-Grimminger D C, Mumat T, Broker T R, Chow L T. Differentiation-dependent up-regulation of the human papillomavirus E7 gene reactivates cellular DNA replication in suprabasal differentiated keratinocytes. Genes Dev. 1995;9:2335–2349. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

