Nucleocapsid and matrix protein contributions to selective human immunodeficiency virus type 1 genomic RNA packaging
- PMID: 9499052
- PMCID: PMC109491
- DOI: 10.1128/JVI.72.3.1983-1993.1998
Nucleocapsid and matrix protein contributions to selective human immunodeficiency virus type 1 genomic RNA packaging
Abstract
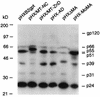
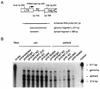
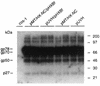
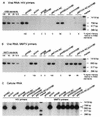
The nucleocapsid protein (NC) of retroviruses plays a major role in genomic RNA packaging, and some evidence has implicated the matrix protein (MA) of certain retroviruses in viral RNA binding. To further investigate the role of NC in the selective recognition of genomic viral RNA and to address the potential contribution of MA in this process, we constructed chimeric and deletion human immunodeficiency virus type 1 (HIV-1) mutants that alter the NC or MA protein. Both HIV and mouse mammary tumor virus (MMTV) NC proteins have two zinc-binding domains and similar basic amino acid compositions but differ substantially in total length, amino acid sequence, and spacing of the zinc-binding motifs. When the entire NC coding sequence of HIV was replaced with the MMTV NC coding sequence, we found that the HIV genome was incorporated into virions at 50% of wild-type levels. Viruses produced from chimeric HIV genomes with complete NC replacements, or with the two NC zinc-binding domains replaced with MMTV sequences, preferentially incorporated HIV genomes when both HIV and MMTV genomes were simultaneously present in the cell. Viruses produced from chimeric MMTV genomes in which the MMTV NC had been replaced with HIV NC preferentially incorporated MMTV genomes when both HIV and MMTV genomes were simultaneously present in the cell. In contrast, viruses produced from chimeric HIV genomes containing the Moloney NC, which contains a single zinc-binding motif, were previously shown to preferentially incorporate Moloney genomic RNA. Taken together, these results indicate that an NC protein with two zinc-binding motifs is required for specific HIV RNA packaging and that the amino acid context of these motifs, while contributing to the process, is less crucial for specificity. The data also suggest that HIV NC may not be the exclusive determinant of RNA selectivity. Analysis of an HIV MA mutant revealed that specific RNA packaging does not require MA protein.
Figures





Similar articles
-
The role of nucleocapsid of HIV-1 in virus assembly.Virology. 1998 Nov 10;251(1):141-57. doi: 10.1006/viro.1998.9374. Virology. 1998. PMID: 9813210
-
Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein.J Virol. 1999 Jul;73(7):5388-401. doi: 10.1128/JVI.73.7.5388-5401.1999. J Virol. 1999. PMID: 10364286 Free PMC article.
-
Single point mutations in the zinc finger motifs of the human immunodeficiency virus type 1 nucleocapsid alter RNA binding specificities of the gag protein and enhance packaging and infectivity.J Virol. 2005 Jun;79(12):7756-67. doi: 10.1128/JVI.79.12.7756-7767.2005. J Virol. 2005. PMID: 15919928 Free PMC article.
-
HIV-1 gag proteins: diverse functions in the virus life cycle.Virology. 1998 Nov 10;251(1):1-15. doi: 10.1006/viro.1998.9398. Virology. 1998. PMID: 9813197 Review.
-
How HIV-1 Gag Manipulates Its Host Cell Proteins: A Focus on Interactors of the Nucleocapsid Domain.Viruses. 2020 Aug 13;12(8):888. doi: 10.3390/v12080888. Viruses. 2020. PMID: 32823718 Free PMC article. Review.
Cited by
-
Yeast three-hybrid screening of rous sarcoma virus mutants with randomly mutagenized minimal packaging signals reveals regions important for gag interactions.J Virol. 2000 Oct;74(19):9167-74. doi: 10.1128/jvi.74.19.9167-9174.2000. J Virol. 2000. PMID: 10982363 Free PMC article.
-
RNA incorporation is critical for retroviral particle integrity after cell membrane assembly of Gag complexes.J Virol. 2002 Dec;76(23):11853-65. doi: 10.1128/jvi.76.23.11853-11865.2002. J Virol. 2002. PMID: 12414928 Free PMC article.
-
Severe acute respiratory syndrome coronavirus nucleocapsid protein confers ability to efficiently produce virus-like particles when substituted for the human immunodeficiency virus nucleocapsid domain.J Biomed Sci. 2008 Nov;15(6):719-29. doi: 10.1007/s11373-008-9265-8. Epub 2008 Jul 1. J Biomed Sci. 2008. PMID: 18592403 Free PMC article.
-
The simian immunodeficiency virus 5' untranslated leader sequence plays a role in intracellular viral protein accumulation and in RNA packaging.J Virol. 2003 Jun;77(11):6284-92. doi: 10.1128/jvi.77.11.6284-6292.2003. J Virol. 2003. PMID: 12743285 Free PMC article.
-
Functional analysis of the murine sarcoma virus RNA packaging sequence.J Virol. 2002 May;76(9):4643-8. doi: 10.1128/jvi.76.9.4643-4648.2002. J Virol. 2002. PMID: 11932430 Free PMC article.
References
-
- Aldovini A, Feinberg M B. Transfection of molecularly cloned HIV genomes. In: Aldovini A, Walker B D, editors. Techniques in HIV research. New York, N.Y: Stockton Press, Inc.; 1990. pp. 147–175.
-
- Aldovini A, Young R A. Construction and analysis of HIV and SIV mutants. Methods Mol Genet. 1994;4A:3–17.
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1987.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

