Association of the human papillomavirus type 11 E1 protein with histone H1
- PMID: 9499053
- PMCID: PMC109492
- DOI: 10.1128/JVI.72.3.1994-2001.1998
Association of the human papillomavirus type 11 E1 protein with histone H1
Abstract
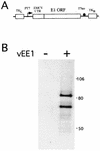
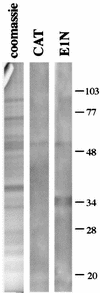
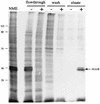
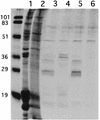
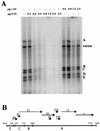
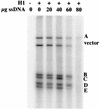
The E1 and E2 proteins are the only virus-encoded factors required for human papillomavirus (HPV) DNA replication. The E1 protein is a DNA helicase responsible for initiation of DNA replication at the viral origin. Its recruitment to the origin is facilitated by binding to E2, for which specific recognition elements are located at the origin. The remaining replication functions for the virus, provided by the host cell's replication machinery, may be mediated by further interactions with E1 and E2. Histone H1 was identified as an HPV type 11 (HPV-11) E1-binding protein by far-Western blotting and by microsequence analyses of a 34-kDa protein purified by E1 affinity chromatography. E1 also bound in vitro to H1 isolated under native conditions in association with intact nucleosomes. In addition, E1 and H1 were coimmunoprecipitated by an E1 antiserum from a nuclear extract prepared from cells expressing recombinant E1. Bound H1 was displaced from HPV-11 DNA by the addition of E1, suggesting that E1 can promote replication initiation and elongation by alteration of viral chromatin structure and disruption of nucleosomes at the replication fork. Furthermore, a region of the HPV-11 genome containing the origin of replication was identified which had weaker affinity for H1 than that of the remaining genome. This result suggests that the presence of a DNA structure at or near the HPV origin facilitates initiation of DNA replication by exclusion of H1. These results are similar to those of studies of simian virus 40 DNA replication, in which a large T antigen-H1 interaction and an H1-resistant region at the origin of DNA replication have also been demonstrated.
Figures

References
-
- Adams C C, Workman J L. Nucleosome displacement in transcription. Cell. 1993;72:305–308. - PubMed
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Berezney R. The nuclear matrix: a heuristic model for investigating genomic organization and function in the cell nucleus. J Cell Biochem. 1991;47:109–123. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

