Functional characterization of naturally occurring variants of human hepatitis B virus containing the core internal deletion mutation
- PMID: 9499073
- PMCID: PMC109512
- DOI: 10.1128/JVI.72.3.2168-2176.1998
Functional characterization of naturally occurring variants of human hepatitis B virus containing the core internal deletion mutation
Abstract
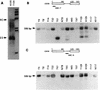
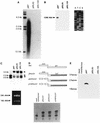
Naturally occurring variants of human hepatitis B virus (HBV) containing the core internal deletion (CID) mutation have been found frequently in HBV carriers worldwide. Despite numerous sequence analysis reports of CID variants in patients, in the past decade, CID variants have not been characterized functionally, and thus their biological significance to HBV infection remains unclear. We report here two different CID variants identified from two patients that are replication defective, most likely due to the absence of detectable core protein. In addition, we were unable to detect the presence of the precore protein and e antigen from CID variants. However, the production of polymerase appeared to be normal. The replication defect of the CID variants can be rescued in trans by complementation with wild-type core protein. The rescued CID variant particles, which utilize the wild-type core protein, presumably are enveloped properly since they can be secreted into the medium and band at a position similar to that of mature wild-type Dane particles, as determined by gradient centrifugation analysis. Our results also provide an explanation for the association of CID variants with helper or wild-type HBV in nature. The significance of CID variants in HBV infection and pathogenesis is discussed.
Figures

Similar articles
-
Out-of-frame versus in-frame core internal deletion variants of human and woodchuck hepatitis B viruses.Virology. 2002 Jan 5;292(1):35-43. doi: 10.1006/viro.2001.1228. Virology. 2002. PMID: 11878906
-
Functional analysis of HBV genomes from patients with fulminant hepatitis.Hepatology. 1998 Nov;28(5):1390-7. doi: 10.1002/hep.510280530. Hepatology. 1998. PMID: 9794926
-
The in vitro replication phenotype of hepatitis B virus (HBV) splice variants Sp3 and Sp9 and their impact on wild-type HBV replication.J Virol. 2024 Apr 16;98(4):e0153823. doi: 10.1128/jvi.01538-23. Epub 2024 Mar 19. J Virol. 2024. PMID: 38501924 Free PMC article.
-
Hepatitis B virus pre-S/S variants in liver diseases.World J Gastroenterol. 2018 Apr 14;24(14):1507-1520. doi: 10.3748/wjg.v24.i14.1507. World J Gastroenterol. 2018. PMID: 29662289 Free PMC article. Review.
-
Virion Secretion of Hepatitis B Virus Naturally Occurring Core Antigen Variants.Cells. 2020 Dec 30;10(1):43. doi: 10.3390/cells10010043. Cells. 2020. PMID: 33396864 Free PMC article. Review.
Cited by
-
Low pathogenic avian influenza virus isolates with different levels of defective genome segments vary in pathogenicity and transmission efficiency.Vet Res. 2020 Aug 28;51(1):108. doi: 10.1186/s13567-020-00833-6. Vet Res. 2020. PMID: 32859269 Free PMC article.
-
Increased expression of Gp96 by HBx-induced NF-κB activation feedback enhances hepatitis B virus production.PLoS One. 2013 Jun 11;8(6):e65588. doi: 10.1371/journal.pone.0065588. Print 2013. PLoS One. 2013. PMID: 23776506 Free PMC article.
-
Hepatitis B virus capsid assembly is enhanced by naturally occurring mutation F97L.J Virol. 2004 Sep;78(17):9538-43. doi: 10.1128/JVI.78.17.9538-9543.2004. J Virol. 2004. PMID: 15308745 Free PMC article.
-
Comparison of complete sequences of hepatitis B virus genotype C between inactive carriers and hepatocellular carcinoma patients before and after seroconversion.J Gastroenterol. 2007 Oct;42(10):837-44. doi: 10.1007/s00535-007-2100-6. Epub 2007 Oct 15. J Gastroenterol. 2007. PMID: 17940837
-
Low-level secretion of human hepatitis B virus virions caused by two independent, naturally occurring mutations (P5T and L60V) in the capsid protein.J Virol. 2000 Oct;74(19):9099-105. doi: 10.1128/jvi.74.19.9099-9105.2000. J Virol. 2000. PMID: 10982356 Free PMC article.
References
-
- Ackrill A M, Naoumov N V, Eddleston A L W F, Williams R. Comparison of pre-core/core hepatitis B virus region in liver tissue and serum from patients with chronic hepatitis B infection. J Hepatol. 1992;16:224–227. - PubMed
-
- Ackrill A M, Naoumov N V, Eddleston A L W F, Williams R. Specific deletions in the hepatitis B virus core open reading frame in patients with chronic active hepatitis. Br J Med Virol. 1993;41:165–169. - PubMed
-
- Akarca U S, Lok A S F. Naturally occurring core-gene-defective hepatitis B viruses. J Gen Virol. 1995;76:1821–1826. - PubMed
-
- Aye T T, Uchida T, Becker S O, Hirashima M, Shikata T, Komine F, Moriyama M, Arakawa Y, Mima S, Mizokami M, Lau J Y N. Variations of hepatitis B virus precore/core gene sequence in acute and fulminant hepatitis B. Dig Dis Sci. 1994;39:1281–1287. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

