Antisense downregulation of N-myc1 in woodchuck hepatoma cells reverses the malignant phenotype
- PMID: 9499076
- PMCID: PMC109515
- DOI: 10.1128/JVI.72.3.2192-2198.1998
Antisense downregulation of N-myc1 in woodchuck hepatoma cells reverses the malignant phenotype
Abstract
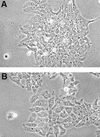
Cell line WH44KA is a highly malignant woodchuck hepatoma cell line. WH44KA cells contain a single woodchuck hepatitis virus (WHV) DNA integration in the 3' untranslated region of exon 3 of the woodchuck N-myc1 gene. The highly rearranged WHV DNA contains WHV enhancers which activate the N-myc promoter, and a hybrid N-myc1-WHV mRNA is produced, which leads to a high steady-state level of N-myc1 protein. To investigate whether continuous N-myc1 expression is required to maintain the tumor phenotype, we knocked out N-myc expression using a WHV-N-myc1 antisense vector. We identified two WH44KA antisense cell lines, designated 4-5 and 4-11, in which steady-state N-mycl protein levels were reduced by 95 and 80%, respectively. The growth rates of both antisense cell lines were reduced in comparison to those of wild-type and vector controls. The phenotype of 4-5 and 4-11 cells changed to a flattened appearance, and the cells exhibited contact inhibition. Colony-forming ability in soft agar was reduced by 92% for 4-5 cells and by 88% for 4-11 cells. Cell line 4-11 formed only small, slow-growing tumors in nude mice, consistent with a low level of N-myc1 remaining in the cells. In contrast, 4-5 cells, in which N-myc protein was reduced by greater than 95%, failed to form tumors in nude mice. The integrated WHV DNA contained the complete WHV X gene (WHx) and its promoter; however, we did not detect any WHx protein in the cells by using a sensitive assay. These data demonstrate that N-myc overexpression is required to maintain the malignant phenotype of WH44KA woodchuck hepatoma cells and provide a direct function for integrated WHV DNA in hepatocarcinogenesis.
Figures





Similar articles
-
Recurrence of WHV integration in the b3n locus in woodchuck hepatocellular carcinoma.Virology. 1995 Dec 1;214(1):229-34. doi: 10.1006/viro.1995.9936. Virology. 1995. PMID: 8525620
-
Integration of woodchuck hepatitis and N-myc rearrangement determine size and histologic grade of hepatic tumors.Hepatology. 2004 Apr;39(4):1008-16. doi: 10.1002/hep.20106. Hepatology. 2004. PMID: 15057905
-
Woodchuck hepatitis virus DNA integration in a common chromosomal region of the woodchuck genome in two independent hepatocellular carcinomas.Arch Virol. 1997;142(3):499-509. doi: 10.1007/s007050050096. Arch Virol. 1997. PMID: 9349296
-
Woodchuck hepatitis virus enhancer I and enhancer II are both involved in N-myc2 activation in woodchuck liver tumors.J Virol. 1998 Jul;72(7):6175-80. doi: 10.1128/JVI.72.7.6175-6180.1998. J Virol. 1998. PMID: 9621085 Free PMC article.
-
The woodchuck: an animal model for hepatitis B virus infection in man.Intervirology. 1995;38(1-2):100-12. doi: 10.1159/000150418. Intervirology. 1995. PMID: 8666518 Review.
Cited by
-
Interaction of the hepatitis B spliced protein with cathepsin B promotes hepatoma cell migration and invasion.J Virol. 2012 Dec;86(24):13533-41. doi: 10.1128/JVI.02095-12. Epub 2012 Oct 3. J Virol. 2012. PMID: 23035214 Free PMC article.
-
Dr.VIS: a database of human disease-related viral integration sites.Nucleic Acids Res. 2012 Jan;40(Database issue):D1041-6. doi: 10.1093/nar/gkr1142. Epub 2011 Dec 1. Nucleic Acids Res. 2012. PMID: 22135288 Free PMC article.
-
Viruses associated with human cancer.Biochim Biophys Acta. 2008 Mar;1782(3):127-50. doi: 10.1016/j.bbadis.2007.12.005. Epub 2007 Dec 23. Biochim Biophys Acta. 2008. PMID: 18201576 Free PMC article. Review.
References
-
- Beasley R P, Lin C C, Hwang L Y, Chien C S. Hepatocellular carcinomas and hepatitis B virus: a prospective study of 22,707 men in Taiwan. Lancet. 1981;72:1129–1133. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

