The gene product of human cytomegalovirus open reading frame UL56 binds the pac motif and has specific nuclease activity
- PMID: 9499084
- PMCID: PMC109523
- DOI: 10.1128/JVI.72.3.2259-2264.1998
The gene product of human cytomegalovirus open reading frame UL56 binds the pac motif and has specific nuclease activity
Abstract
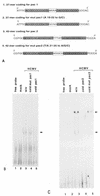
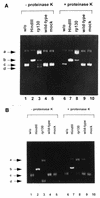
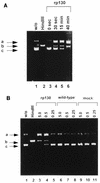
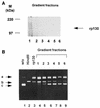
Using the cis-acting human cytomegalovirus (HCMV) packaging elements (pac 1 and pac 2) as DNA probes, specific DNA-protein complexes were detected by electrophoretic mobility shift assay (EMSA) in both HCMV-infected cell nuclear extracts and recombinant baculovirus-infected cell extracts containing the HCMV p130 (pUL56) protein. DNA-binding proteins, which were common in uninfected and infected cell extracts, were also detected. Mutational analysis showed that only the AT-rich core sequences in these cis-acting motifs, 5'-TAAAAA-3' (pac 1) and 5'-TTTTAT-3' (pac 2), were required for specific DNA-protein complex formation. The specificity of the DNA-protein complexes was confirmed by EMSA competition. Furthermore, a specific endonuclease activity was found to be associated with lysates of baculovirus-infected cells expressing recombinant p130 (rp130). This nuclease activity was time dependent, related to the amount of rp130 in the assay, and ATP independent. Nuclease activity remained associated with rp130 after partial purification by sucrose gradient centrifugation, suggesting that this activity is a property of HCMV p130. We propose a possible involvement of p130 in HCMV DNA packaging.
Figures


References
-
- Addison C, Rixon F J, Preston V G. Herpes simplex virus type 1 UL28 gene product is important for the formation of mature capsids. J Virol. 1990;71:2377–2384. - PubMed
-
- Alford C A, Britt W J. Cytomegalovirus. In: Roizman B, Whiteley R J, Lopez C, et al., editors. The human herpesviruses. New York, N.Y: Raven Press, Ltd.; 1993. pp. 227–255.
-
- Bogner E, Reschke M, Reis B, Reis E, Britt W, Radsak K. Recognition of compartmentalized intracellular analogs of glycoprotein H of human cytomegalovirus. Arch Virol. 1992;126:67–80. - PubMed
-
- Bogner E, Reschke M, Reis B, Mockenhaupt T, Radsak K. Identification of the gene product encoded by ORF UL56 of human cytomegalovirus genome. Virology. 1993;196:290–293. - PubMed
-
- Chee M, Bankier A, Beck S, Bohni R, Brown C, Cerny R, Horsnell T, Huchison C, Kouzarides T, Martignetti E, Preddie E, Satchnell S, Tomlinson P, Weston K, Barell B. Analysis of the protein coding content of the sequence of human cytomegalovirus AD169. Curr Top Microbiol Immunol. 1990;154:125–169. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

