CD4 glycoprotein degradation induced by human immunodeficiency virus type 1 Vpu protein requires the function of proteasomes and the ubiquitin-conjugating pathway
- PMID: 9499087
- PMCID: PMC109526
- DOI: 10.1128/JVI.72.3.2280-2288.1998
CD4 glycoprotein degradation induced by human immunodeficiency virus type 1 Vpu protein requires the function of proteasomes and the ubiquitin-conjugating pathway
Abstract
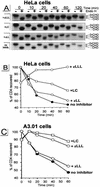
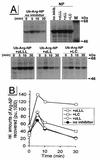
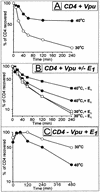
The human immunodeficiency virus type 1 (HIV-1) vpu gene encodes a type I anchored integral membrane phosphoprotein with two independent functions. First, it regulates virus release from a post-endoplasmic reticulum (ER) compartment by an ion channel activity mediated by its transmembrane anchor. Second, it induces the selective down regulation of host cell receptor proteins (CD4 and major histocompatibility complex class I molecules) in a process involving its phosphorylated cytoplasmic tail. In the present work, we show that the Vpu-induced proteolysis of nascent CD4 can be completely blocked by peptide aldehydes that act as competitive inhibitors of proteasome function and also by lactacystin, which blocks proteasome activity by covalently binding to the catalytic beta subunits of proteasomes. The sensitivity of Vpu-induced CD4 degradation to proteasome inhibitors paralleled the inhibition of proteasome degradation of a model ubiquitinated substrate. Characterization of CD4-associated oligosaccharides indicated that CD4 rescued from Vpu-induced degradation by proteasome inhibitors is exported from the ER to the Golgi complex. This finding suggests that retranslocation of CD4 from the ER to the cytosol may be coupled to its proteasomal degradation. CD4 degradation mediated by Vpu does not require the ER chaperone calnexin and is dependent on an intact ubiquitin-conjugating system. This was demonstrated by inhibition of CD4 degradation (i) in cells expressing a thermally inactivated form of the ubiquitin-activating enzyme E1 or (ii) following expression of a mutant form of ubiquitin (Lys48 mutated to Arg48) known to compromise ubiquitin targeting by interfering with the formation of polyubiquitin complexes. CD4 degradation was also prevented by altering the four Lys residues in its cytosolic domain to Arg, suggesting a role for ubiquitination of one or more of these residues in the process of degradation. The results clearly demonstrate a role for the cytosolic ubiquitin-proteasome pathway in the process of Vpu-induced CD4 degradation. In contrast to other viral proteins (human cytomegalovirus US2 and US11), however, whose translocation of host ER molecules into the cytosol occurs in the presence of proteasome inhibitors, Vpu-targeted CD4 remains in the ER in a transport-competent form when proteasome activity is blocked.
Figures



References
-
- Aiken C J, Konner J, Landau N R, Lenburg E, Trono D. Nef induces CD4 endocytosis: requirement of a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell. 1994;76:503–513. - PubMed
-
- Bergeron J J M, Brenner M B, Thomas D Y, Williams D B. Calnexin: a membrane bound chaperone of the endoplasmic reticulum. Trends Biochem Sci. 1994;19:124–128. - PubMed
-
- Bonifacino J S, Klausner R D. Degradation of proteins in the endoplasmic reticulum. In: Ciechanover A, Schwartz A L, editors. Cellular proteolytic systems. Vol. 15. New York, N.Y: Wiley-Liss; 1994. pp. 137–160.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

