The R33 G protein-coupled receptor gene of rat cytomegalovirus plays an essential role in the pathogenesis of viral infection
- PMID: 9499096
- PMCID: PMC109535
- DOI: 10.1128/JVI.72.3.2352-2363.1998
The R33 G protein-coupled receptor gene of rat cytomegalovirus plays an essential role in the pathogenesis of viral infection
Abstract
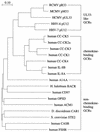
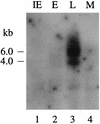
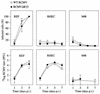
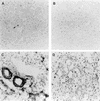
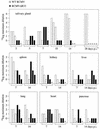
We have identified a rat cytomegalovirus (RCMV) gene that encodes a G-protein-coupled receptor (GCR) homolog. This gene (R33) belongs to a family that includes the human cytomegalovirus UL33 gene. R33 was found to be transcribed during the late phase of RCMV infection in rat embryo fibroblasts. Unlike the mRNAs from all the other members of the UL33 family that have been studied to date, the R33 mRNA is not spliced. To study the function of the R33 gene, we constructed an RCMV strain in which the R33 open reading frame is disrupted. The mutant strain (RCMV deltaR33) did not show differences in replication from wild-type RCMV upon infection of several rat cell types in vitro. However, marked differences were seen between the mutant and wild-type strain in the pathogenesis of infection in immunocompromised rats. First, the mutant strain induced a significantly lower mortality than the wild-type virus did. Second, in contrast to wild-type RCMV, the mutant strain did not efficiently replicate in the salivary gland epithelial cells of immunocompromised rats. Although viral DNA was detected in salivary glands of RCMV deltaR33-infected rats up to 14 days postinfection, it could not be detected at later time points. This indicates that although the strain with R33 deleted is probably transported to the salivary glands in a similar fashion to that for wild-type virus, the mutant virus is not able to either enter or replicate in salivary gland epithelial cells. We conclude that the RCMV R33 gene plays a vital role in the pathogenesis of infection.
Figures




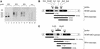

References
-
- Ahuja S K, Murphy P M. Molecular piracy of mammalian interleukin-8 receptor type B by herpesvirus saimiri. J Biol Chem. 1993;268:20691–20694. - PubMed
-
- Aida K, Koishi S, Tawata M, Onaya T. Molecular cloning of a putative Ca(2+)-sensing receptor cDNA from human kidney. Biochem Biophys Res Commun. 1995;214:524–529. - PubMed
-
- Arvatakis L, Geras-Raaka E, Varma A, Gershengorn M C, Cesarman E. Human herpesvirus KSHV encodes a constitutively active G-protein-coupled receptor linked to cell proliferation. Nature. 1997;385:347–350. - PubMed
-
- Baer R J, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatful G F, Hudson G S, Satchwell S C, Sequin C, Tuffnell P S, Barrell B G. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

