Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo
- PMID: 9499399
- PMCID: PMC316582
- DOI: 10.1101/gad.12.5.627
Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo
Abstract
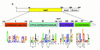
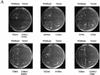
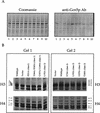
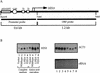
Gcn5p is a transcriptional coactivator required for correct expression of various genes in yeast. Several transcriptional regulators, including Gcn5p, possess intrinsic histone acetyltransferase (HAT) activity in vitro. However, whether the HAT activity of any of these proteins is required for gene activation remains unclear. Here, we demonstrate that the HAT activity of Gcn5p is critical for transcriptional activation of target genes in vivo. Core histones are hyperacetylated in cells overproducing functional Gcn5p, and promoters of Gcn5p-regulated genes are associated with hyperacetylated histones upon activation by low-copy Gcn5p. Point mutations within the Gcn5p catalytic domain abolish both promoter-directed histone acetylation and Gcn5p-mediated transcriptional activation. These data provide the first in vivo evidence that promoter-specific histone acetylation, catalyzed by functional Gcn5p, plays a critical role in gene activation.
Figures





References
-
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In: Altman R, Brutlag D, Karp P, Lathrop R, Searls D, editors. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. Menlo Park, CA: AAAI Press; 1994. pp. 28–36. - PubMed
-
- Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. - PubMed
-
- Barlev NA, Candau R, Wang L, Darpino P, Silverman N, Berger SL. Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J Biol Chem. 1995;270:19337–19344. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
