The crystal structure of Pyrococcus furiosus ornithine carbamoyltransferase reveals a key role for oligomerization in enzyme stability at extremely high temperatures
- PMID: 9501170
- PMCID: PMC19649
- DOI: 10.1073/pnas.95.6.2801
The crystal structure of Pyrococcus furiosus ornithine carbamoyltransferase reveals a key role for oligomerization in enzyme stability at extremely high temperatures
Abstract
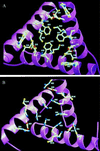
The Pyrococcus furiosus (PF) ornithine carbamoyltransferase (OTCase; EC 2.1.3.3) is an extremely heat-stable enzyme that maintains about 50% of its activity after heat treatment for 60 min at 100 degrees C. To understand the molecular basis of thermostability of this enzyme, we have determined its three-dimensional structure at a resolution of 2.7 A and compared it with the previously reported structures of OTCases isolated from mesophilic bacteria. Most OTCases investigated up to now are homotrimeric and devoid of allosteric properties. A striking exception is the catabolic OTCase from Pseudomonas aeruginosa, which is allosterically regulated and built up of four trimers disposed in a tetrahedral manner, an architecture that actually underlies the allostery of the enzyme. We now report that the thermostable PF OTCase (420 kDa) presents the same 23-point group symmetry. The enzyme displays Michaelis-Menten kinetics. A detailed comparison of the two enzymes suggests that, in OTCases, not only allostery but also thermophily was achieved through oligomerization of a trimer as a common catalytic motif. Thermal stabilization of the PF OTCase dodecamer is mainly the result of hydrophobic interfaces between trimers, at positions where allosteric binding sites have been identified in the allosteric enzyme. The present crystallographic analysis of PF OTCase provides a structural illustration that oligomerization can play a major role in extreme thermal stabilization.
Figures


References
-
- Stetter K O. FEMS Microbiol Rev. 1996;18:149–158. - PubMed
-
- Glansdorff N. In: Cellular and Molecular Biology. Neidhardt F, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 408–433.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

