Detection of Clostridium proteoclasticum and closely related strains in the rumen by competitive PCR
- PMID: 9501430
- PMCID: PMC106345
- DOI: 10.1128/AEM.64.3.907-913.1998
Detection of Clostridium proteoclasticum and closely related strains in the rumen by competitive PCR
Abstract
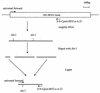
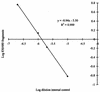
A competitive PCR technique was used to enumerate the proteolytic bacterium Clostridium proteoclasticum from the rumen. A PCR primer, which circumscribes this organism and several closely related strains, was designed for a variable region within their 16S rRNA genes and was used in conjunction with a universal forward primer. This primer pair was tested for specificity against 85 ruminal bacterial strains. An internal control DNA was constructed for use in competitive PCRs and was shown to amplify under the same reaction conditions and with the same amplification efficiency as the target DNA. DNA from a known number of C. proteoclasticum cells was coamplified with the internal control to construct a standard curve. Rumen samples were collected from eight dairy cows fed four diets in rotation: high nitrogen, high nitrogen supplemented with carbohydrate, low nitrogen, and low nitrogen supplemented with carbohydrate. DNA extracted from these and spiked with internal control DNA was amplified with the C. proteoclasticum primer pair. The relative intensities of the PCR products were used to quantitate the numbers of C. proteoclasticum cell equivalents from the rumen samples. The numbers ranged from 2.01 x 10(6) ml-1 to 3.12 x 10(7) ml-1. There was no significant effect on the numbers of C. proteoclasticum detected in rumen samples among cows fed the four diets. The utility of the competitive PCR approach for quantifying ruminal bacterial populations in vivo and the occurrence of C. proteoclasticum in forage-fed dairy cows are discussed.
Figures




References
-
- Attwood G T, Reilly K. Identification of proteolytic rumen bacteria isolated from New Zealand cattle. J Appl Bacteriol. 1995;79:22–29. - PubMed
-
- Attwood G T, Reilly K. Characterization of proteolytic activities of rumen bacterial isolates from forage-fed cattle. J Appl Bacteriol. 1996;81:545–552. - PubMed
-
- Attwood G T, Reilly K, Patel B K C. Clostridium proteoclasticum sp. nov., a novel proteolytic bacterium from the bovine rumen. Int J Syst Bacteriol. 1996;46:753–758. - PubMed
-
- Bej A K, Mahbubani M H, Atlas R M. Amplification of nucleic acids by polymerase chain reaction (PCR) and other methods and their applications. Crit Rev Biochem Mol Biol. 1991;26:301–334. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

