Biochemical and genetic characterization of an extracellular protease from Pseudomonas fluorescens CY091
- PMID: 9501431
- PMCID: PMC106346
- DOI: 10.1128/AEM.64.3.914-921.1998
Biochemical and genetic characterization of an extracellular protease from Pseudomonas fluorescens CY091
Abstract
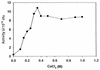
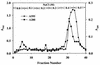
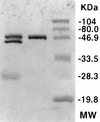
Pseudomonas fluorescens CY091 cultures produce an extracellular protease with an estimated molecular mass of 50 kDa. Production of this enzyme (designated AprX) was observed in media containing CaCl2 or SrCl2 but not in media containing ZnCl2, MgCl2, or MnCl2. The requirement of Ca2+ (or Sr2+) for enzyme production was concentration dependent, and the optimal concentration for production was determined to be 0.35 mM. Following ammonium sulfate precipitation and ion-exchange chromatography, the AprX in the culture supernatant was purified to near electrophoretic homogeneity. Over 20% of the enzyme activity was retained in the AprX sample which had been heated in boiling water for 10 min, indicating that the enzyme is highly resistant to heat inactivation. The enzyme activity was almost completely inhibited in the presence of 1 mM 1,10-phenanthroline, but only 30% of the activity was inhibited in the presence of 1 mM EGTA. The gene encoding AprX was cloned from the genome of P. fluorescens CY091 by isolating cosmid clones capable of restoring the protease production in a nonproteolytic mutant of strain CY091. The genomic region of strain CY091 containing the aprX gene was located within a 7.3-kb DNA fragment. Analysis of the complete nucleotide sequence of this 7.3-kb fragment revealed the presence of a cluster of genes required for the production of extracellular AprX in P. fluorescens and Escherichia coli. The AprX protein showed 50 to 60% identity in amino acid sequence to the related proteases produced by Pseudomonas aeruginosa and Erwinia chrysanthemi. Two conserved sequence domains possibly associated with Ca2+ and Zn2+ binding were identified. Immediately adjacent to the aprX structural gene, a gene (inh) encoding a putative protease inhibitor and three genes (aprD, aprE, and aprF), possibly required for the transport of AprX, were also identified. The organization of the gene cluster involved in the synthesis and secretion of AprX in P. fluorescens CY091 appears to be somewhat different from that previously demonstrated in P. aeruginosa and E. chrysanthemi.
Figures





References
-
- Alichanidis E, Andrews A T. Some properties of the extracellular protease produced by the psychrotrophic bacterium Pseudomonas fluorescens strain AR-11. Biochim Biophys Acta. 1977;485:424–433. - PubMed
-
- Bond J S, Butler P E. Intracellular proteases. Annu Rev Biochem. 1987;56:333–364. - PubMed
-
- Bradford M M. A rapid and sensitive method for quantitation of microgram quantities of proteins utilizing protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
-
- Delepelaire P, Wandersman C. Protease secretion by Erwinia chrysanthemi. J Biol Chem. 1989;264:9083–9089. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

