Bacterial community dynamics during start-up of a trickle-bed bioreactor degrading aromatic compounds
- PMID: 9501433
- PMCID: PMC106348
- DOI: 10.1128/AEM.64.3.930-939.1998
Bacterial community dynamics during start-up of a trickle-bed bioreactor degrading aromatic compounds
Abstract
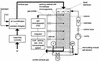
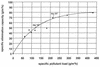
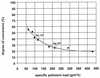
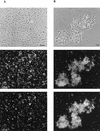
This study was performed with a laboratory-scale fixed-bed bioreactor degrading a mixture of aromatic compounds (Solvesso100). The starter culture for the bioreactor was prepared in a fermentor with a wastewater sample of a care painting facility as the inoculum and Solvesso100 as the sole carbon source. The bacterial community dynamics in the fermentor and the bioreactor were examined by a conventional isolation procedure and in situ hybridization with fluorescently labeled rRNA-targeted oligonucleotides. Two significant shifts in the bacterial community structure could be demonstrated. The original inoculum from the wastewater of the car factory was rich in proteobacteria of the alpha and beta subclasses, while the final fermentor enrichment was dominated by bacteria closely related to Pseudomonas putida or Pseudomonas mendocina, which both belong to the gamma subclass of the class Proteobacteria. A second significant shift was observed when the fermentor culture was transferred as inoculum to the trickle-bed bioreactor. The community structure in the bioreactor gradually returned to a higher complexity, with the dominance of beta and alpha subclass proteobacteria, whereas the gamma subclass proteobacteria sharply declined. Obviously, the preceded pollutant adaptant did not lead to a significant enrichment of bacteria that finally dominated in the trickle-bed bioreactor. In the course of experiments, three new 16S as well as 23S rRNA-targeted probes for beta subclass proteobacteria were designed, probe SUBU1237 for the genera Burkholderia and Sutterella, probe ALBO34a for the genera Alcaligenes and Bordetella, and probe Bcv13b for Burkholderia cepacia and Burkholderia vietnamiensis. Bacteria hybridizing with the probe Bcv13b represented the main Solvesso100-degrading population in the reactor.
Figures


References
-
- Amann R I, Ludwig W. Typing in situ with probes. In: Priest F G, et al., editors. Bacterial diversity and systematics. New York, N.Y: Plenum Press; 1994. pp. 115–135.
-
- Amann R I, Ludwig W, Schulze R, Spring S, Moore E, Schleifer K-H. rRNA-targeted oligonucleotide probes for the identification of genuine and former pseudomonads. Syst Appl Microbiol. 1996;19:501–509.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

