PCR amplification of ribosomal DNA for species identification in the plant pathogen genus Phytophthora
- PMID: 9501434
- PMCID: PMC106350
- DOI: 10.1128/AEM.64.3.948-954.1998
PCR amplification of ribosomal DNA for species identification in the plant pathogen genus Phytophthora
Abstract
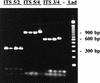
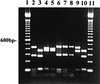
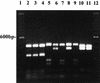
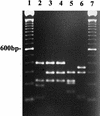
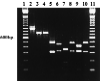
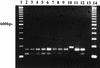
We have developed a PCR procedure to amplify DNA for quick identification of the economically important species from each of the six taxonomic groups in the plant pathogen genus Phytophthora. This procedure involves amplification of the 5.8S ribosomal DNA gene and internal transcribed spacers (ITS) with the ITS primers ITS 5 and ITS 4. Restriction digests of the amplified DNA products were conducted with the restriction enzymes RsaI, MspI, and HaeIII. Restriction fragment patterns were similar after digestions with RsaI for the following species: P. capsici and P. citricola; P. infestans, P. cactorum, and P. mirabilis; P. fragariae, P. cinnamomi, and P. megasperma from peach; P. palmivora, P. citrophthora, P. erythroseptica, and P. cryptogea; and P. megasperma from raspberry and P. sojae. Restriction digests with MspI separated P. capsici from P. citricola and separated P. cactorum from P. infestans and P. mirabilis. Restriction digests with HaeIII separated P. citrophthora from P. cryptogea, P. cinnamomi from P. fragariae and P. megasperma on peach, P. palmivora from P. citrophthora, and P. megasperma on raspberry from P. sojae. P. infestans and P. mirabilis digests were identical and P. cryptogea and P. erythroseptica digests were identical with all restriction enzymes tested. A unique DNA sequence from the ITS region I in P. capsici was used to develop a primer called PCAP. The PCAP primer was used in PCRs with ITS 1 and amplified only isolates of P. capsici, P. citricola, and P. citrophthora and not 13 other species in the genus. Restriction digests with MspI separated P. capsici from the other two species. PCR was superior to traditional isolation methods for detection of P. capsici in infected bell pepper tissue in field samples. The techniques described will provide a powerful tool for identification of the major species in the genus Phytophthora.
Figures

References
-
- Benson D M. Detection of Phytophthora cinnamomi in azalea with commercial serological assay kits. Plant Dis. 1991;75:478–482.
-
- Bruns T D, White T J, Taylor J W. Fungal molecular systematics. Annu Rev Ecol Syst. 1991;22:525–564.
-
- Cooke D E L, Duncan J M. Phylogenetic analysis of Phytophthora species based on ITS 1 and 2 sequences of the rDNA gene repeat. Mycol Res. 1997;101:667–677.
-
- Duncan J. Fungal and bacterial diseases. Dundee, Scotland: Report of the Scottish Crops research Institute; 1995.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

