Altered srf expression in Bacillus subtilis resulting from changes in culture pH is dependent on the Spo0K oligopeptide permease and the ComQX system of extracellular control
- PMID: 9515911
- PMCID: PMC107042
- DOI: 10.1128/JB.180.6.1438-1445.1998
Altered srf expression in Bacillus subtilis resulting from changes in culture pH is dependent on the Spo0K oligopeptide permease and the ComQX system of extracellular control
Abstract
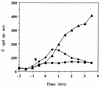
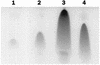
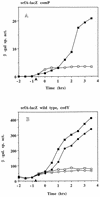
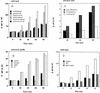
The expression of the srf operon of Bacillus subtilis, encoding surfactin synthetase and the competence regulatory protein ComS, was observed to be reduced when cells were grown in a rich glucose- and glutamine-containing medium in which late-growth culture pH was 5.0 or lower. The production of the surfactin synthetase subunits and of surfactin itself was also reduced. Raising the pH to near neutrality resulted in dramatic increases in srf expression and surfactin production. This apparent pH-dependent induction of srf expression required spo0K, which encodes the oligopeptide permease that functions in cell-density-dependent control of sporulation and competence, but not CSF, the competence-inducing pheromone that regulates srf expression in a Spo0K-dependent manner. Both ComP and ComA, the two-component regulatory pair that stimulates cell-density-dependent srf transcription, were required for optimal expression of srf at low and high pHs, but ComP was not required for pH-dependent srf induction. The known negative regulators of srf, RapC and CodY, were found not to function significantly in pH-dependent srf expression. Late-growth culture supernatants at low pH were not active in inducing srf expression in cells of low-density cultures but were rendered active when their pH was raised to near neutrality. ComQ (and very likely the srf-inducing pheromone ComX) and Spo0K were found to be required for the extracellular induction of srf-lacZ at neutral pH. The results suggest that srf expression, in response to changes in culture pH, requires Spo0K and another, as yet unidentified, extracellular factor. The study also provides evidence consistent with the hypothesis that ComP acts both positively and negatively in the regulation of ComA and that both activities are controlled by the ComX pheromone.
Figures



Similar articles
-
Purification and characterization of an extracellular peptide factor that affects two different developmental pathways in Bacillus subtilis.Genes Dev. 1996 Aug 15;10(16):2014-24. doi: 10.1101/gad.10.16.2014. Genes Dev. 1996. PMID: 8769645
-
Plasmid-amplified comS enhances genetic competence and suppresses sinR in Bacillus subtilis.J Bacteriol. 1996 Sep;178(17):5144-52. doi: 10.1128/jb.178.17.5144-5152.1996. J Bacteriol. 1996. PMID: 8752331 Free PMC article.
-
Convergent sensing pathways mediate response to two extracellular competence factors in Bacillus subtilis.Genes Dev. 1995 Mar 1;9(5):547-58. doi: 10.1101/gad.9.5.547. Genes Dev. 1995. PMID: 7698645
-
Regulation of peptide antibiotic production in Bacillus.Mol Microbiol. 1993 Mar;7(5):631-6. doi: 10.1111/j.1365-2958.1993.tb01154.x. Mol Microbiol. 1993. PMID: 7682277 Review.
-
Research advances in the identification of regulatory mechanisms of surfactin production by Bacillus: a review.Microb Cell Fact. 2024 Apr 2;23(1):100. doi: 10.1186/s12934-024-02372-7. Microb Cell Fact. 2024. PMID: 38566071 Free PMC article. Review.
Cited by
-
Modulation of the ComA-dependent quorum response in Bacillus subtilis by multiple Rap proteins and Phr peptides.J Bacteriol. 2006 Jul;188(14):5273-85. doi: 10.1128/JB.00300-06. J Bacteriol. 2006. PMID: 16816200 Free PMC article.
-
An overview and future prospects of recombinant protein production in Bacillus subtilis.Appl Microbiol Biotechnol. 2021 Sep;105(18):6607-6626. doi: 10.1007/s00253-021-11533-2. Epub 2021 Sep 1. Appl Microbiol Biotechnol. 2021. PMID: 34468804 Review.
-
Stepwise modifications of genetic parts reinforce the secretory production of nattokinase in Bacillus subtilis.Microb Biotechnol. 2018 Sep;11(5):930-942. doi: 10.1111/1751-7915.13298. Epub 2018 Jul 8. Microb Biotechnol. 2018. PMID: 29984489 Free PMC article.
-
Extracellular DNA release by undomesticated Bacillus subtilis is regulated by early competence.PLoS One. 2012;7(11):e48716. doi: 10.1371/journal.pone.0048716. Epub 2012 Nov 2. PLoS One. 2012. PMID: 23133654 Free PMC article.
-
Overview of the Antimicrobial Compounds Produced by Members of the Bacillus subtilis Group.Front Microbiol. 2019 Feb 26;10:302. doi: 10.3389/fmicb.2019.00302. eCollection 2019. Front Microbiol. 2019. PMID: 30873135 Free PMC article. Review.
References
-
- Arima K, Kainuma A, Tamura G. Surfactin, a crystalline peptide lipid surfactant produced by Bacillus subtilis: isolation, characterization, and its inhibition of fibrin clot formation. Biochem Biophys Res Commun. 1968;31:488–494. - PubMed
-
- Bernheimer A W, Avegad L S. Nature and properties of a cytolytic agent produced by Bacillus subtilis. J Gen Microbiol. 1970;61:361–369. - PubMed
-
- Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
-
- Cosmina P, Rodriguez F, de Ferra F, Grandi G, Perego M, Venema G, van Sinderen D. Sequence and analysis of the genetic locus responsible for surfactin synthesis in Bacillus subtilis. Mol Microbiol. 1993;8:821–831. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

