Activation of Escherichia coli rRNA transcription by FIS during a growth cycle
- PMID: 9515922
- PMCID: PMC107053
- DOI: 10.1128/JB.180.6.1525-1532.1998
Activation of Escherichia coli rRNA transcription by FIS during a growth cycle
Abstract
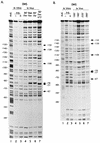
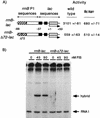
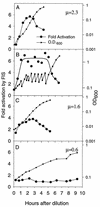
rRNA transcription in Escherichia coli is activated by the FIS protein, which binds upstream of rrnp1 promoters and interacts directly with RNA polymerase. Analysis of the contribution of FIS to rrn transcription under changing physiological conditions is complicated by several factors: the wide variation in cellular FIS concentrations with growth conditions, the contributions of several other regulatory systems to rRNA synthesis, and the pleiotropy of fis mutations. In this report, we show by in vivo footprinting and Western blot analysis that occupancy of the rrnBp1 FIS sites correlates with cellular levels of FIS. We find, using two methods of measurement (pulse induction of a FIS-activated hybrid promoter and primer extension from an unstable transcript made from rrnBp1), that the extent of transcription activation by FIS parallels the degree of FIS site occupancy and therefore cellular FIS levels. FIS activates transcription throughout exponential growth at low culture density, but rrnp1 transcription increases independently of FIS immediately following upshift, before FIS accumulates. These results support the model that FIS is one of a set of overlapping signals that together contribute to transcription from rrnp1 promoters during steady-state growth.
Figures
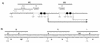

References
-
- Appleman, J. A., and R. L. Gourse. Unpublished results.
-
- Bachmann B J. Linkage map of Escherichia coli K-12, edition 7. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 807–876.
-
- Bartlett, M., T. Gaal, W. Ross, and R. L. Gourse. RNA polymerase mutants defective in the NTP-sensing mechanism for growth rate-dependent control of rRNA transcription. Submitted for publication.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

