The folate branch of the methionine biosynthesis pathway in Streptomyces lividans: disruption of the 5,10-methylenetetrahydrofolate reductase gene leads to methionine auxotrophy
- PMID: 9515933
- PMCID: PMC107064
- DOI: 10.1128/JB.180.6.1586-1591.1998
The folate branch of the methionine biosynthesis pathway in Streptomyces lividans: disruption of the 5,10-methylenetetrahydrofolate reductase gene leads to methionine auxotrophy
Abstract
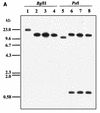
In enterobacteria, the methyl group of methionine is donated by 5-methyltetrahydrofolate that is synthesized from N5,10-methylenetetrahydrofolate by the 5,10-methylenetetrahydrofolate reductase. The Streptomyces lividans metF gene, which encodes 5,10-methylenetetrahydrofolate reductase, has been cloned. It encodes a protein of 307 amino acids with a deduced molecular mass of 33,271 Da. S1 exonuclease mapping of the transcription initiation site showed that the metF gene is expressed, forming a leaderless mRNA. A 13-bp tandem repeat located immediately upstream of the promoter region shows homology with the consensus MetR-binding sequence of Salmonella typhimurium. Expression of metF in multicopy plasmids in S. lividans resulted in accumulation of a 32-kDa protein, as shown by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Disruption of the metF gene led to methionine auxotrophy. Integration of the disrupting plasmid at the metF locus was confirmed by Southern hybridization in three randomly isolated transformants. The methionine auxotrophy was complemented by transformation of the auxotrophs with an undisrupted metF gene. These results indicate that the folate branch is essential for methionine biosynthesis in streptomycetes, as occurs in enterobacteria.
Figures


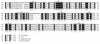



References
-
- Anné J, Van Mellaert L. Streptomyces lividans as host for heterologous protein production. FEMS Microbiol Lett. 1993;114:121–128. - PubMed
-
- Belfaiza J, Parsot C, Martel A, Buothier de la Tour C, Margarita D, Cohen G N, Saint-Girons I. Evolution in the biosynthesis pathways: two enzymes catalyzing consecutive steps in methionine biosynthesis originate from a common ancestor and possess a similar regulatory region. Proc Natl Acad Sci USA. 1986;83:867–871. - PMC - PubMed
-
- Bibb M J, Cohen S N. Gene expression in Streptomyces: construction and application of promoter-probe plasmid vectors in Streptomyces lividans. Mol Gen Genet. 1982;187:265–277. - PubMed
-
- Chary, V. K., and J. F. Martín. Unpublished data.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

