A single side chain prevents Escherichia coli DNA polymerase I (Klenow fragment) from incorporating ribonucleotides
- PMID: 9520378
- PMCID: PMC19848
- DOI: 10.1073/pnas.95.7.3402
A single side chain prevents Escherichia coli DNA polymerase I (Klenow fragment) from incorporating ribonucleotides
Abstract
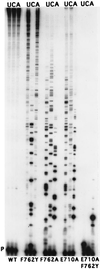
Although nucleic acid polymerases from different families show striking similarities in structure, they maintain stringent specificity for the sugar structure of the incoming nucleoside triphosphate. The Klenow fragment of E. coli DNA polymerase I selects its natural substrates, deoxynucleotides, over ribonucleotides by several thousand fold. Analysis of mutant Klenow fragment derivatives indicates that discrimination is provided by the Glu-710 side chain which sterically blocks the 2'-OH of an incoming rNTP. A nearby aromatic side chain, at position 762, plays an important role in constraining the nucleotide so that the Glu-710 "steric gate" can be fully effective. Even with the E710A mutation, which is extremely permissive for addition of a single ribonucleotide to a DNA primer, Klenow fragment does not efficiently synthesize pure RNA, indicating that additional barriers prevent the incorporation of successive ribonucleotides.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

