Parallel analysis of genetic selections using whole genome oligonucleotide arrays
- PMID: 9520439
- PMCID: PMC19909
- DOI: 10.1073/pnas.95.7.3752
Parallel analysis of genetic selections using whole genome oligonucleotide arrays
Abstract
Thousands of genes have recently been sequenced in organisms ranging from Escherichia coli to human. For the majority of these genes, however, available sequence does not define a biological role. Efficient functional characterization of these genes requires strategies for scaling genetic analyses to the whole genome level. Plasmid-based library selections are an established approach to the functional analysis of uncharacterized genes and can help elucidate biological function by identifying, for example, physical interactors for a gene and genetic enhancers and suppressors of mutant phenotypes. The application of these selections to every gene in a eukaryotic genome, however, is generally limited by the need to manipulate and sequence hundreds of DNA plasmids. We present an alternative approach in which identification of nucleic acids is accomplished by direct hybridization to high-density oligonucleotide arrays. Based on the complete sequence of Saccharomyces cerevisiae, high-density arrays containing oligonucleotides complementary to every gene in the yeast genome have been designed and synthesized. Two-hybrid protein-protein interaction screens were carried out for S. cerevisiae genes implicated in mRNA splicing and microtubule assembly. Hybridization of labeled DNA derived from positive clones is sufficient to characterize the results of a screen in a single experiment, allowing rapid determination of both established and previously unknown biological interactions. These results demonstrate the use of oligonucleotide arrays for the analysis of two-hybrid screens. This approach should be generally applicable to the analysis of a range of genetic selections.
Figures
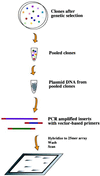
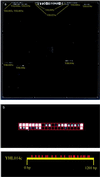
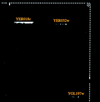
References
-
- Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, Feldmann H, Galibert F, Hoheisel J D, Jacq C, Johnston M, et al. Science. 1996;274:563–567. - PubMed
-
- Oliver S G. Nature (London) 1996;379:597–600. - PubMed
-
- Fields S. Nat Genet. 1997;15:325–327. - PubMed
-
- Fromont-Racine M, Rain J C, Legrain P. Nat Genet. 1997;16:277–282. - PubMed
-
- Hoffman C S, Winston F. Gene. 1987;57:267–272. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

