Unusual features of the Drosophila melanogaster telomere transposable element HeT-A are conserved in Drosophila yakuba telomere elements
- PMID: 9520442
- PMCID: PMC19912
- DOI: 10.1073/pnas.95.7.3770
Unusual features of the Drosophila melanogaster telomere transposable element HeT-A are conserved in Drosophila yakuba telomere elements
Abstract
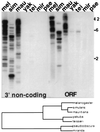
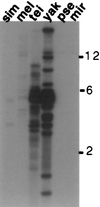
HeT-A was the first transposable element shown to have a bona fide role in chromosome structure, maintenance of telomeres in Drosophila melanogaster. HeT-A has hallmarks of non-long-terminal-repeat (non-LTR) retrotransposable elements but also has several unique features. We have now isolated HeT-A elements from Drosophila yakuba, showing that the retrotransposon mechanism of telomere maintenance predates the separation of D. melanogaster and D. yakuba (5-15 million years ago). HeT-A elements from the two species show significant sequence divergence, yet unusual features seen in HeT-Amel are conserved in HeT-Ayak. In both species, HeT-A elements are found in head-to-tail tandem arrays in telomeric heterochromatin. In both species, nearly half of the HeT-A sequence is noncoding and shows a distinctive imperfect repeat pattern of A-rich segments. Neither element encodes reverse transcriptase. The HeT-Amel promoter appears to be intermediate between the promoters of non-LTR and of LTR retrotransposons. The HeT-Ayak promoter shows similar features. HeT-Amel has a frameshift within the coding region. HeT-Ayak does not require a frameshift but shows conservation of the polypeptide sequence of the frameshifted product of D. melanogaster.
Figures
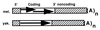


References
-
- Henderson E. In: Telomeres. Greider C, Blackburn E, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. pp. 11–34.
-
- Pardue M-L. In: Telomeres. Greider C, Blackburn E, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. pp. 339–370.
-
- Pardue M-L, Danilevskaya O N, Lowenhaupt K, Slot F, Traverse K L. Trends Genet. 1996;12:48–52. - PubMed
-
- Danilevskaya O N, Petrov D A, Pavlova M A, Koga A, Kurenova E V, Hartl D L. Chromosoma. 1992;102:32–40. - PubMed
-
- Biessmann H, Kasravi B, Bui T, Fujiwara G, Champion L E, Mason J M. Chromosoma. 1994;103:90–98. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

