BioViews: Java-based tools for genomic data visualization
- PMID: 9521932
- PMCID: PMC310693
- DOI: 10.1101/gr.8.3.291
BioViews: Java-based tools for genomic data visualization
Abstract
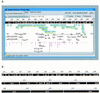
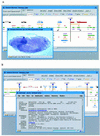
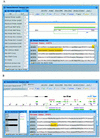
Visualization tools for bioinformatics ideally should provide universal access to the most current data in an interactive and intuitive graphical user interface. Since the introduction of Java, a language designed for distributed programming over the Web, the technology now exists to build a genomic data visualization tool that meets these requirements. Using Java we have developed a prototype genome browser applet (BioViews) that incorporates a three-level graphical view of genomic data: a physical map, an annotated sequence map, and a DNA sequence display. Annotated biological features are displayed on the physical and sequence-based maps, and the different views are interconnected. The applet is linked to several databases and can retrieve features and display hyperlinked textual data on selected features. In addition to browsing genomic data, different types of analyses can be performed interactively and the results of these analyses visualized alongside prior annotations. Our genome browser is built on top of extensible, reusable graphic components specifically designed for bioinformatics. Other groups can (and do) reuse this work in various ways. Genome centers can reuse large parts of the genome browser with minor modifications, bioinformatics groups working on sequence analysis can reuse components to build front ends for analysis programs, and biology laboratories can reuse components to publish results as dynamic Web documents.
Figures


References
-
- Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Altschul SF, Boguski MS, Gish W, Wootton JC. Issues in searching molecular sequence databases. Nature Genet. 1994;6:119–129. - PubMed
-
- Bederson B, Hollan J. User Interface Software and Technology. New York, NY: ACM Press; 1994. >Pad++: A zooming graphical interface for exploring alternate interface physics.
-
- Bier EA, Stone MC, Pier K, Buxton W, DeRose TD. Proceedings of SIGGRAPH ’93, Computer Graphics Annual Conference Series. New York, NY: ACM Press; 1993. Toolglass and magic lenses: The see-through interface; pp. 73–80.
-
- Dunham I, Durbin R, Thierry-Mieg J, Bentley DR. Physical mapping projects and ACeDB. In: Bishop MJ, editor. Guide to human genome computing. New York, NY: Harcourt Brace; 1994. pp. 110–158.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
