Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites
- PMID: 9525604
- PMCID: PMC109729
- DOI: 10.1128/JVI.72.4.2846-2854.1998
Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites
Abstract
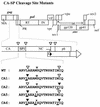
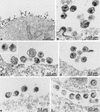
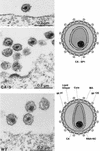
Retroviruses are produced as immature particles containing structural polyproteins, which are subsequently cleaved by the viral proteinase (PR). Extracellular maturation leads to condensation of the spherical core to a capsid shell formed by the capsid (CA) protein, which encases the genomic RNA complexed with nucleocapsid (NC) proteins. CA and NC are separated by a short spacer peptide (spacer peptide 1 [SP1]) on the human immunodeficiency virus type 1 (HIV-1) Gag polyprotein and released by sequential PR-mediated cleavages. To assess the role of individual cleavages in maturation, we constructed point mutations abolishing cleavage at these sites, either alone or in combination. When all three sites between CA and NC were mutated, immature particles containing stable CA-NC were observed, with no apparent effect on other cleavages. Delayed maturation with irregular morphology of the ribonucleoprotein core was observed when cleavage of SP1 from NC was prevented. Blocking the release of SP1 from CA, on the other hand, yielded normal condensation of the ribonucleoprotein core but prevented capsid condensation. A thin, electron-dense layer near the viral membrane was observed in this case, and mutant capsids were significantly less stable against detergent treatment than wild-type HIV-1. We suggest that HIV maturation is a sequential process controlled by the rate of cleavage at individual sites. Initial rapid cleavage at the C terminus of SP1 releases the RNA-binding NC protein and leads to condensation of the ribonucleoprotein core. Subsequently, CA is separated from the membrane by cleavage between the matrix protein and CA, and late release of SP1 from CA is required for capsid condensation.
Figures

References
-
- Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

