Adenovirus preterminal protein binds to the CAD enzyme at active sites of viral DNA replication on the nuclear matrix
- PMID: 9525610
- PMCID: PMC109735
- DOI: 10.1128/JVI.72.4.2896-2904.1998
Adenovirus preterminal protein binds to the CAD enzyme at active sites of viral DNA replication on the nuclear matrix
Abstract
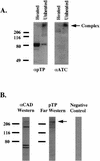
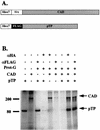
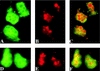
Adenovirus (Ad) replicative complexes form at discrete sites on the nuclear matrix (NM) via an interaction mediated by the precursor of the terminal protein (pTP). The identities of cellular proteins involved in these complexes have remained obscure. We present evidence that pTP binds to a multifunctional pyrimidine biosynthesis enzyme found at replication domains on the NM. Far-Western blotting identified proteins of 150 and 240 kDa that had pTP binding activity. Amino acid sequencing of the 150-kDa band revealed sequence identity to carbamyl phosphate synthetase I (CPS I) and a high degree of homology to the related trifunctional enzyme known as CAD (for carbamyl phosphate synthetase, aspartate transcarbamylase, and dihydroorotase). Western blotting with an antibody directed against CAD detected a 240-kDa band that comigrated with that detected by pTP far-Western blotting. Binding experiments showed that a pTP-CAD complex was immunoprecipitable from cell extracts in which pTP was expressed by a vaccinia virus recombinant. Additionally, in vitro-translated epitope-tagged pTP and CAD were immunoprecipitable as a complex, indicating the occurrence of a protein-protein interaction. Confocal fluorescence microscopy of Ad-infected NM showed that pTP and CAD colocalized in nuclear foci. Both pTP and CAD were confirmed to colocalize with active sites of replication detected by bromodeoxyuridine incorporation. These data support the concept that the pTP-CAD interaction may allow anchorage of Ad replication complexes in the proximity of required cellular factors and may help to segregate replicated and unreplicated viral DNA.
Figures





References
-
- Ausubell F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons Inc.; 1995.
-
- Berezney R, Coffey D S. Identification of a nuclear protein matrix. Biochem Biophys Res Commun. 1974;60:1410–1417. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

