Functional analysis of the human immunodeficiency virus type 1 Rev protein oligomerization interface
- PMID: 9525614
- PMCID: PMC109739
- DOI: 10.1128/JVI.72.4.2935-2944.1998
Functional analysis of the human immunodeficiency virus type 1 Rev protein oligomerization interface
Abstract
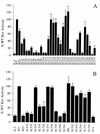
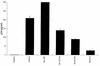
The expression of human immunodeficiency virus type 1 (HIV-1) structural proteins requires the action of the viral trans-regulatory protein Rev. Rev is a nuclear shuttle protein that directly binds to its cis-acting Rev response element (RRE) RNA target sequence. Subsequent oligomerization of Rev monomers on the RRE and interaction of Rev with a cellular cofactor(s) result in the cytoplasmic accumulation of RRE-containing viral mRNAs. Moreover, Rev by itself is exported from the nucleus to the cytoplasm. Although it has been demonstrated that Rev multimerization is critically required for Rev activity and hence for HIV-1 replication, the number of Rev monomers required to form a trans-activation-competent complex on the RRE is unknown. Here we report a systematic analysis of the putative multimerization domains within the Rev trans-activator protein. We identify the amino acid residues which are part of the proposed single hydrophobic surface patch in the Rev amino terminus that mediates intermolecular interactions. Furthermore, we show that the expression of a multimerization-deficient Rev mutant blocks HIV-1 replication in a trans-dominant (dominant-negative) fashion.
Figures



References
-
- Auer M, Gremlich H-U, Seifert J-M, Daly T J, Parslow T G, Casari G, Gstach H. Helix-loop-helix motif in HIV-1 Rev. Biochem. 1994;33:2988–2996. - PubMed
-
- Bartel D P, Zapp M L, Green M R, Szostak J W. HIV-1 Rev regulation involves recognition of non-Watson-Crick base pairs in viral RNA. Cell. 1991;67:529–536. - PubMed
-
- Berger J, Aepinus C, Dobrovnik M, Fleckenstein B, Hauber J, Böhnlein E. Mutational analysis of functional domains in the HIV-1 Rev trans-regulatory protein. Virology. 1991;183:630–635. - PubMed
-
- Berger J, Hauber J, Hauber R, Geiger R, Cullen B R. Secreted placental alkaline phosphatase: a powerful new quantitative indicator of gene expression in eukaryotic cells. Gene. 1988;66:1–10. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

