Adenovirus E1B 55K represses p53 activation in vitro
- PMID: 9525640
- PMCID: PMC109770
- DOI: 10.1128/JVI.72.4.3146-3154.1998
Adenovirus E1B 55K represses p53 activation in vitro
Abstract
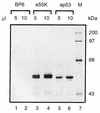
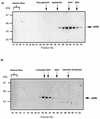
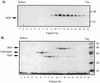
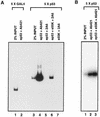
Adenovirus E1B 55K protein cooperates with E1A gene products to induce cell transformation. E1B 55K mediates its effects by binding to and inhibiting the transcriptional activation and growth-suppression functions of the tumor suppressor p53. Previous studies in vivo have suggested that E1B 55K has an active role in repressing p53 transcriptional activation and that this repression function is directed to specific promoters through E1B 55K's interaction with DNA-bound p53. Flag-tagged E1B 55K (e55K) was expressed with the baculovirus expression system and immunopurified. Gel filtration, velocity sedimentation centrifugation, and glutaraldehyde cross-linking indicated that e55K is a dimer with a nonglobular conformation. e55K bound directly to purified p53, causing an approximately 10-fold increase in p53 affinity for tandem binding sites. Using in vitro transcription assays reconstituted with purified p53, e55K, and HeLa cell nuclear extracts, we found that e55K specifically repressed p53 activation. These results demonstrate that as postulated from earlier transient expression experiments, E1B 55K is a specific repressor of transcription from a promoter with bound p53. Since HeLa nuclear extracts contain little detectable histone protein, E1B 55K probably represses transcription through direct or indirect interactions with the RNA polymerase II transcription machinery.
Figures




References
-
- Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. - PubMed
-
- Auble D T, Hansen K E, Mueller C G, Lane W S, Thorner J, Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. - PubMed
-
- Ayer D E, Kretzner L, Eisenman R N. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993;72:211–222. - PubMed
-
- Buckbinder L, Talbott R, Velasco-Miguel S, Takenaka I, Faha B, Seizinger B R, Kley N. Induction of the growth inhibitor IGF-binding protein 3 by p53. Nature. 1995;377:646–649. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

