Cleavage of human immunodeficiency virus type 1 proteinase from the N-terminally adjacent p6* protein is essential for efficient Gag polyprotein processing and viral infectivity
- PMID: 9525682
- PMCID: PMC109854
- DOI: 10.1128/JVI.72.4.3459-3463.1998
Cleavage of human immunodeficiency virus type 1 proteinase from the N-terminally adjacent p6* protein is essential for efficient Gag polyprotein processing and viral infectivity
Abstract
Maturation of infectious human immunodeficiency virus (HIV) particles requires proteolytic cleavage of the structural polyproteins by the viral proteinase (PR), which is itself encoded as part of the Gag-Pol polyprotein. Expression of truncated PR-containing sequences in heterologous systems has mostly led to the autocatalytic release of an 11-kDa species of PR which is capable of processing all known cleavage sites on the viral precursor proteins. Relatively little is known about cleavages within the nascent virus particle, on the other hand, and controversial results concerning the active PR species inside the virion and the relative activities of extended PR species have been reported. Here, we report that HIV type 1 (HIV-1) particles of four different strains obtained from different cell lines contain an 11-kDa PR, with no extended PR proteins detectable. Furthermore, mutation of the N-terminal PR cleavage site leading to production of an N-terminally extended 17-kDa PR species caused a severe defect in Gag polyprotein processing and a complete loss of viral infectivity. We conclude that N-terminal release of PR from the HIV-1 polyprotein is essential for viral replication and suggest that extended versions of PR may have a transient function in the proteolytic cascade.
Figures


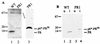
References
-
- Beissinger M, Paulus C, Bayer P, Wolf H, Rösch P, Wagner R. Sequence-specific resonance assignments of the 1H-NMR spectra and structural characterization in solution of the HIV-1 transframe protein p6*. Eur J Biochem. 1996;237:383–392. - PubMed
-
- Co E, Koelsch G, Lin Y, Ido E, Hartsuck J A, Tang J. Proteolytic processing mechanisms of a miniprecursor of the aspartic proteinase of human immunodeficiency virus type 1. Biochemistry. 1994;33:1248–1254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

