Requirement for end-joining and checkpoint functions, but not RAD52-mediated recombination, after EcoRI endonuclease cleavage of Saccharomyces cerevisiae DNA
- PMID: 9528760
- PMCID: PMC121418
- DOI: 10.1128/MCB.18.4.1891
Requirement for end-joining and checkpoint functions, but not RAD52-mediated recombination, after EcoRI endonuclease cleavage of Saccharomyces cerevisiae DNA
Abstract
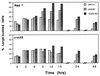
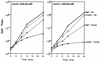
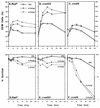
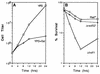
RAD52 and RAD9 are required for the repair of double-strand breaks (DSBs) induced by physical and chemical DNA-damaging agents in Saccharomyces cerevisiae. Analysis of EcoRI endonuclease expression in vivo revealed that, in contrast to DSBs containing damaged or modified termini, chromosomal DSBs retaining complementary ends could be repaired in rad52 mutants and in G1-phase Rad+ cells. Continuous EcoRI-induced scission of chromosomal DNA blocked the growth of rad52 mutants, with most cells arrested in G2 phase. Surprisingly, rad52 mutants were not more sensitive to EcoRI-induced cell killing than wild-type strains. In contrast, endonuclease expression was lethal in cells deficient in Ku-mediated end joining. Checkpoint-defective rad9 mutants did not arrest cell cycling and lost viability rapidly when EcoRI was expressed. Synthesis of the endonuclease produced extensive breakage of nuclear DNA and stimulated interchromosomal recombination. These results and those of additional experiments indicate that cohesive ended DSBs in chromosomal DNA can be accurately repaired by RAD52-mediated recombination and by recombination-independent complementary end joining in yeast cells.
Figures




References
-
- Bai Y, Symington L S. A RAD52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 1996;10:2025–2037. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
