The microsatellite sequence (CT)n x (GA)n promotes stable chromosomal integration of large tandem arrays of functional human U2 small nuclear RNA genes
- PMID: 9528797
- PMCID: PMC121475
- DOI: 10.1128/MCB.18.4.2262
The microsatellite sequence (CT)n x (GA)n promotes stable chromosomal integration of large tandem arrays of functional human U2 small nuclear RNA genes
Abstract
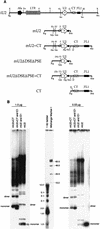
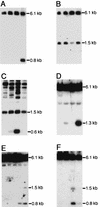
The multigene family encoding human U2 small nuclear RNA (snRNA) is organized as a single large tandem array containing 5 to 25 copies of a 6.1-kb repeat unit (the RNU2 locus). Remarkably, each of the repeat units within an individual U2 tandem array appears to be identical except for an irregular dinucleotide tract, known as the CT microsatellite, which exhibits minor length and sequence polymorphism. Using a somatic cell genetic assay, we previously noticed that the CT microsatellite appeared to stabilize artificial tandem arrays of U2 snRNA genes. We now demonstrate that the CT microsatellite is required to establish large tandem arrays of transcriptionally active U2 genes, increasing both the average and maximum size of the resulting arrays. In contrast, the CT microsatellite has no effect on the average or maximal size of artificial arrays containing transcriptionally inactive U2 genes that lack key promoter elements. Our data reinforce the connection between recombination and transcription. Active U2 transcription interferes with establishment or maintenance of the U2 tandem array, and the CT microsatellite opposes these effects, perhaps by binding GAGA or GAGA-related factors which alter local chromatin structure. We speculate that the mechanisms responsible for maintenance of tandem arrays containing active promoters may differ from those that maintain tandem arrays of transcriptionally inactive sequences.
Figures




References
-
- Ares M, Jr, Chung J-S, Giglio L, Weiner A M. Distinct factors with Sp1 and NF-A specificities bind to adjacent functional elements of the human U2 snRNA gene enhancer. Genes Dev. 1987;1:808–817. - PubMed
-
- Armour J A, Crosier M, Malcolm S, Chan J C, Jeffreys A J. Human minisatellite loci composed of interspersed GGA-GGT triplet repeats. Proc R Soc Lond Ser B. 1995;261:345–349. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
