Requirement for both Shc and phosphatidylinositol 3' kinase signaling pathways in polyomavirus middle T-mediated mammary tumorigenesis
- PMID: 9528804
- PMCID: PMC121489
- DOI: 10.1128/MCB.18.4.2344
Requirement for both Shc and phosphatidylinositol 3' kinase signaling pathways in polyomavirus middle T-mediated mammary tumorigenesis
Abstract
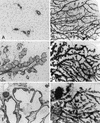
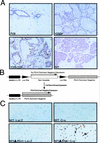
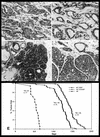
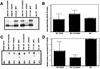
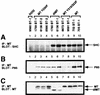
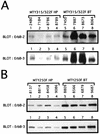
Transgenic mice expressing the polyomavirus (PyV) middle T antigen (MT) develop multifocal mammary tumors which frequently metastasize to the lung. The potent transforming activity of PyV MT is correlated with its capacity to activate and associate with a number of signaling molecules, including the Src family tyrosine kinases, the 85-kDa Src homology 2 subunit of the phosphatidylinositol 3' (PI-3') kinase, and the Shc adapter protein. To uncover the role of these signaling proteins in MT-mediated mammary tumorigenesis, we have generated transgenic mice that express mutant PyV MT antigens decoupled from either the Shc or the PI-3' kinase signaling pathway. In contrast to the rapid induction of metastatic mammary tumors observed in the strains expressing wild-type PyV MT, mammary epithelial cell-specific expression of either mutant PyV MT resulted in the induction of extensive mammary epithelial hyperplasias. The mammary epithelial hyperplasias expressing the mutant PyV MT defective in recruiting the PI-3' kinase were highly apoptotic, suggesting that recruitment of PI-3' kinase by MT affects cell survival. Whereas the initial phenotypes observed in both strains were global mammary epithelial hyperplasias, focal mammary tumors eventually arose in all female transgenic mice. Genetic and biochemical analyses of tumorigenesis in the transgenic strains expressing the PyV MT mutant lacking the Shc binding site revealed that a proportion of the metastatic tumors arising in these mice displayed evidence of reversion of the mutant Shc binding site. In contrast, no evidence of reversion of the PI-3' kinase binding site was noted in tumors derived from the strains expressing the PI-3' kinase binding site MT mutant. Tumor progression in both mutant strains was further correlated with upregulation of the epidermal growth factor receptor family members which are known to couple to the PI-3' kinase and Shc signaling pathways. Taken together, these observations suggest that PyV MT-mediated tumorigenesis requires activation of both Shc and PI-3' kinase, which appear to be required for stimulation of cell proliferation and survival signaling pathways, respectively.
Figures
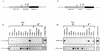

References
-
- Brizuela L, Ulug E T, Jones M A, Courtneidge S A. Induction of interleukin-2 transcription by the hamster polyoma virus middle T antigen: a role for Fyn in T cell signal transduction. Eur J Immunol. 1995;25:385–393. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
