Human matrix attachment regions insulate transgene expression from chromosomal position effects in Drosophila melanogaster
- PMID: 9528807
- PMCID: PMC121496
- DOI: 10.1128/MCB.18.4.2382
Human matrix attachment regions insulate transgene expression from chromosomal position effects in Drosophila melanogaster
Abstract
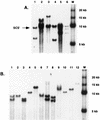
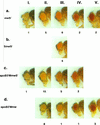
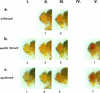
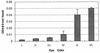
Germ line transformation of white- Drosophila embryos with P-element vectors containing white expression cassettes results in flies with different eye color phenotypes due to position effects at the sites of transgene insertion. These position effects can be cured by specific DNA elements, such as the Drosophila scs and scs' elements, that have insulator activity in vivo. We have used this system to determine whether human matrix attachment regions (MARs) can function as insulator elements in vivo. Two different human MARs, from the apolipoprotein B and alpha1-antitrypsin loci, insulated white transgene expression from position effects in Drosophila melanogaster. Both elements reduced variability in transgene expression without enhancing levels of white gene expression. In contrast, expression of white transgenes containing human DNA segments without matrix-binding activity was highly variable in Drosophila transformants. These data indicate that human MARs can function as insulator elements in vivo.
Figures


References
-
- Bao J J, Reed-Fourquet L, Sifers R N, Kidd V J, Woo S L. Molecular structure and sequence homology of a gene related to α1-antitrypsin in the human genome. Genomics. 1988;2:165–173. - PubMed
-
- Blackhart B D, Ludwig E M, Pierotti V R, Caiati L, Onasch M A, Wallis S C, Powell L, Pease R, Knott T J, Chu M L, et al. Structure of the human apolipoprotein B gene. J Biol Chem. 1986;261:15364–15367. - PubMed
-
- Bode J, Maass K. Chromatin domain surrounding the human interferon-β gene as defined by scaffold-attached regions. Biochemistry. 1988;27:4706–4711. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
