Molecular analysis of Shiga toxigenic Escherichia coli O111:H- proteins which react with sera from patients with hemolytic-uremic syndrome
- PMID: 9529069
- PMCID: PMC108076
- DOI: 10.1128/IAI.66.4.1467-1472.1998
Molecular analysis of Shiga toxigenic Escherichia coli O111:H- proteins which react with sera from patients with hemolytic-uremic syndrome
Abstract
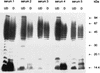
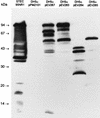
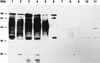
Western blot analysis was used to assess the reactivity of convalescent-phase sera from patients who were associated with an outbreak of hemolytic-uremic syndrome (HUS) caused by fermented sausage contaminated with Shiga toxin-producing Escherichia coli (STEC). The predominant STEC isolated from HUS patients belonged to serotype O111:H-, and reactivity to O111:H- whole-cell lysates, treated or untreated with proteinase K, was examined. As expected, all five serum samples demonstrated a marked anti-lipopolysaccharide response, but several protein bands were also immunoreactive, particularly one with an apparent size of 94 kDa. One convalescent-phase serum sample was subsequently used to screen an O111:H- cosmid bank and 2 of 900 cosmid clones were found to be positive, both of which contained a similar DNA insert. Western blot analysis of one of these clones identified three major immunoreactive protein bands of approximately 94, 70, and 50 kDa. An immune response to the three proteins was detectable with all five convalescent-phase serum samples but not with healthy human serum. Immunoreactive 94- and 50-kDa species were produced by a deletion derivative of the cosmid containing a 7-kb STEC DNA insert. Sequence analysis of this region indicated that it is part of the locus for enterocyte effacement, including the eaeA gene which encodes intimin. The deduced amino acid sequence of the O111:H- intimin was 88.6% identical to intimin from O157:H7 STEC, and the most divergent region was the 200 residues at the carboxyl terminus, which were only 75% identical. Such variation may be antigenically significant as serum from a HUS patient infected only with the O111:H- STEC reacted with intimin from an enteropathogenic E. coli O111 strain, as well as several other eaeA-positive STEC isolates, but not with an eaeA-positive STEC belonging to serotype O157:H-. Sera from two of the other HUS patients also failed to react with intimin from this latter strain. However, intimin from O157:H- STEC did react with serum from a patient infected with both O111:H- and O157:H- STEC.
Figures



References
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Barrett T J, Kaper J B, Jerse A E, Wachsmuth I K. Virulence factors in Shiga-like toxin-producing Escherichia coli isolated from humans and cattle. J Infect Dis. 1992;165:979–980. - PubMed
-
- Beebakhee G, Louie M, De Azavedo J, Brunton J. Cloning and nucleotide sequence of the eae gene homologue from enterohemorrhagic Escherichia coli serotype O157:H7. FEMS Microbiol Lett. 1992;91:63–68. - PubMed
-
- Donnenberg M S, Kaper J B, Finlay B B. Interactions between enteropathogenic Escherichia coli and host epithelial cells. Trends Microbiol. 1997;5:109–114. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

