Identification of a gene involved in assembly of Actinomyces naeslundii T14V type 2 fimbriae
- PMID: 9529071
- PMCID: PMC108078
- DOI: 10.1128/IAI.66.4.1482-1491.1998
Identification of a gene involved in assembly of Actinomyces naeslundii T14V type 2 fimbriae
Abstract
The nucleotide sequence of the Actinomyces naeslundii T14V type 2 fimbrial structural subunit gene, fimA, and the 3' flanking DNA region was determined. The fimA gene encoded a 535-amino-acid precursor subunit protein (FimA) which included both N-terminal leader and C-terminal cell wall sorting sequences. A second gene, designated orf365, that encoded a 365-amino-acid protein which contained a putative transmembrane segment was identified immediately 3' to fimA. Mutants in which either fimA or orf365 was replaced with a kanamycin resistance gene did not participate in type 2 fimbriae-mediated coaggregation with Streptococcus oralis 34. Type 2 fimbrial antigen was not detected in cell extracts of the fimA mutant by Western blotting with anti-A. naeslundii type 2 fimbrial antibody, but the subunit protein was detected in extracts of the orf365 mutant. The subunit protein detected in this mutant also was immunostained by an antibody raised against a synthetic peptide representing the C-terminal 20 amino acid residues of the predicted FimA. The antipeptide antibody reacted with FimA isolated from the recombinant Escherichia coli clone containing fimA but did not react with purified type 2 fimbriae in extracts of the wild-type strain. These results indicate that synthesis of type 2 fimbriae in A. naeslundii T14V may involve posttranslational cleavage of both the N-terminal and C-terminal peptides of the precursor subunit and also the expression of orf365.
Figures




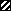 , fimA;
, fimA;
 , orf365. The chromosomal region where plasmid
integration occurred as mediated by the Campbell insertion-duplication
mechanism is also indicated (▧).
, orf365. The chromosomal region where plasmid
integration occurred as mediated by the Campbell insertion-duplication
mechanism is also indicated (▧).
References
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Campbell A M. Episomes. Adv Genet. 1962;11:101–145.
-
- Cisar J O. Fimbrial lectins of the oral actinomyces. In: Mirelman D, editor. Microbial lectins and agglutinins: properties and biological activity. New York, N.Y: John Wiley & Sons, Inc.; 1986. pp. 183–196.
-
- Cisar J O, Barsumian E L, Curl S H, Vatter A E, Sandberg A L, Siraganian R P. Detection and localization of a lectin on Actinomyces viscosusT14V by monoclonal antibodies. J Immunol. 1981;127:1318–1322. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

