A new member of the S-layer protein family: characterization of the crs gene from Campylobacter rectus
- PMID: 9529076
- PMCID: PMC108083
- DOI: 10.1128/IAI.66.4.1521-1526.1998
A new member of the S-layer protein family: characterization of the crs gene from Campylobacter rectus
Abstract
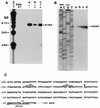
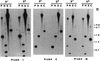
Strains of the periodontal pathogen Campylobacter rectus express a 150- to 166-kDa protein on their cell surface. This protein forms a paracrystalline lattice, called the surface layer (S-layer), on the outer membrane of this gram-negative bacterium. To initiate a genetic analysis of the function of the S-layer in the pathogenesis of C. rectus, we have cloned and characterized its gene. The S-layer gene (crs) from C. rectus 314 encodes a cell surface protein which does not have a cleaved signal peptide at its amino terminus. Although the amino acid sequence deduced from the crs gene has 50% identity with the amino-terminal 30 amino acids of the four S-layer proteins from Campylobacter fetus, the similarity decreases to less than 16% over the rest of the protein. Thus, the crs gene from C. rectus encodes a novel S-layer protein whose precise role in pathogenesis may differ from that of S-layer proteins from other organisms. Southern and Northern blot analyses with probes from different segments of the crs gene indicate that the S-layer gene is a single-copy, monocistronic gene in C. rectus. RNA end mapping and sequence analyses were used to define the crs promoter; there is an exact match to the Escherichia coli -10 promoter consensus sequence but only a weak match to the -35 consensus element. Southern blots of DNA from another strain of C. rectus, ATCC 33238, demonstrated that the crs gene is also present in that strain but that there are numerous restriction fragment length polymorphisms in the second half of the gene. This finding suggests that the carboxy halves of the S-layer proteins from strains 314 and 33238 differ. It remains to be determined whether the diversities in sequence are reflected in functional or antigenic differences important for the pathogenesis of different C. rectus isolates.
Figures




Similar articles
-
Campylobacter surface-layers (S-layers) and immune evasion.Ann Periodontol. 2002 Dec;7(1):43-53. doi: 10.1902/annals.2002.7.1.43. Ann Periodontol. 2002. PMID: 16013216 Free PMC article. Review.
-
Cloning and characterization of two bistructural S-layer-RTX proteins from Campylobacter rectus.J Bacteriol. 1999 Apr;181(8):2501-6. doi: 10.1128/JB.181.8.2501-2506.1999. J Bacteriol. 1999. PMID: 10198015 Free PMC article.
-
Purification and characterization of Campylobacter rectus surface layer proteins.Infect Immun. 1997 Feb;65(2):478-83. doi: 10.1128/iai.65.2.478-483.1997. Infect Immun. 1997. PMID: 9009300 Free PMC article.
-
The S-layer protein from Campylobacter rectus: sequence determination and function of the recombinant protein.FEMS Microbiol Lett. 1998 Sep 15;166(2):275-81. doi: 10.1111/j.1574-6968.1998.tb13901.x. FEMS Microbiol Lett. 1998. PMID: 9770285
-
Role of the S-layer proteins of Campylobacter fetus in serum-resistance and antigenic variation: a model of bacterial pathogenesis.Am J Med Sci. 1993 Nov;306(5):325-9. doi: 10.1097/00000441-199311000-00011. Am J Med Sci. 1993. PMID: 8238090 Review.
Cited by
-
Campylobacter surface-layers (S-layers) and immune evasion.Ann Periodontol. 2002 Dec;7(1):43-53. doi: 10.1902/annals.2002.7.1.43. Ann Periodontol. 2002. PMID: 16013216 Free PMC article. Review.
-
The role of ATP-binding cassette transporters in bacterial pathogenicity.Protoplasma. 2012 Oct;249(4):919-42. doi: 10.1007/s00709-011-0360-8. Epub 2012 Jan 13. Protoplasma. 2012. PMID: 22246051 Review.
-
RTX proteins: a highly diverse family secreted by a common mechanism.FEMS Microbiol Rev. 2010 Nov;34(6):1076-112. doi: 10.1111/j.1574-6976.2010.00231.x. FEMS Microbiol Rev. 2010. PMID: 20528947 Free PMC article. Review.
-
Campylobacter fetus surface layer proteins are transported by a type I secretion system.J Bacteriol. 1998 Dec;180(24):6450-8. doi: 10.1128/JB.180.24.6450-6458.1998. J Bacteriol. 1998. PMID: 9851986 Free PMC article.
-
Cloning and characterization of two bistructural S-layer-RTX proteins from Campylobacter rectus.J Bacteriol. 1999 Apr;181(8):2501-6. doi: 10.1128/JB.181.8.2501-2506.1999. J Bacteriol. 1999. PMID: 10198015 Free PMC article.
References
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1994.
-
- Blaser M J. Role of the S-layer proteins of Campylobacter fetus in serum-resistance and antigenic variation: a model of bacterial pathogenesis. Am J Med Sci. 1994;306:325–329. - PubMed
-
- Blaser M J, Gotschlich E C. Surface array protein of Campylobacter fetus. Cloning and gene structure. J Biol Chem. 1990;265:14529–14535. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

