Maturation of the arginine-specific proteases of Porphyromonas gingivalis W50 is dependent on a functional prR2 protease gene
- PMID: 9529086
- PMCID: PMC108093
- DOI: 10.1128/IAI.66.4.1594-1600.1998
Maturation of the arginine-specific proteases of Porphyromonas gingivalis W50 is dependent on a functional prR2 protease gene
Abstract
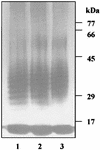
The prpR1 of Porphyromonas gingivalis codes for three distinct enzymes with specificity for arginyl peptide bonds termed RI, RIA, and RIB. These three isoforms comprise the majority of the extracellular, arginine-specific protease activity in P. gingivalis W50. RI is a heterodimer in which the catalytic alpha chain is noncovalently associated with a second chain involved in adherence phenomena. RIA and RIB are both monomeric species. RIA represents the free alpha chain, and RIB is a highly posttranslationally modified form of the alpha chain which is exclusively vesicle or membrane associated and migrates as a diffuse band on sodium dodecyl sulfate-polyacrylamide gel electrophoresis. In previous studies, insertional inactivation of the prpR1 demonstrated that arginine-specific protease activity can also arise from a closely related second gene, prR2. In the present work, the prR2 was insertionally inactivated in P. gingivalis W50 in order to establish the contribution of this locus to the arginine-specific protease activity of this periodontal bacterium. Loss of prR2 function had several effects on prpR1-derived enzymes. First, the total Arg-X activity was reduced by approximately 50% relative to that of the parent strain. The reduction in total activity was a consequence of decreased concentrations of the monomeric enzymes derived from the prpR1, while the heterodimeric enzyme, RI, was unaffected by this mutation. Second, the chromatographic behavior of both the soluble and vesicle- or membrane-associated monomeric enzymes was radically different from the behavior of RIA and RIB from the parent strain. Finally, the vesicle- or membrane-associated enzyme in the prR2 mutant strain lacked the extensive posttranslational additions which are found on RIB in P. gingivalis W50. These data suggest that the product(s) of the prR2 plays a significant role in the maturation pathway of prpR1-derived enzymes, and this may contribute to the coconservation of these two genes in P. gingivalis.
Figures





References
-
- Allaker R P, Aduse-Opoku J, Batten J E, Curtis M A. Natural variation within the principal arginine-specific protease gene, prpR1, of P. gingivalis. Oral Microbiol Immunol. 1997;12:298–302. - PubMed
-
- Curtis M A, Ramakrishnan M, Slaney J M. Characterisation of the trypsin-like enzymes of Porphyromonas gingivalis W83 using a radiolabelled active-site-directed inhibitor. J Gen Microbiol. 1993;139:949–955. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

