Asexual sporulation in Aspergillus nidulans
- PMID: 9529886
- PMCID: PMC98905
- DOI: 10.1128/MMBR.62.1.35-54.1998
Asexual sporulation in Aspergillus nidulans
Erratum in
- Microbiol Mol Biol Rev 1998 Jun;62(2):545
Abstract
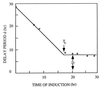
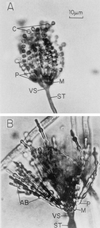
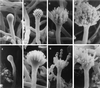
The formation of mitotically derived spores, called conidia, is a common reproductive mode in filamentous fungi, particularly among the large fungal class Ascomycetes. Asexual sporulation strategies are nearly as varied as fungal species; however, the formation of conidiophores, specialized multicellular reproductive structures, by the filamentous fungus Aspergillus nidulans has emerged as the leading model for understanding the mechanisms that control fungal sporulation. Initiation of A. nidulans conidiophore formation can occur either as a programmed event in the life cycle in response to intrinsic signals or to environmental stresses such as nutrient deprivation. In either case, a development-specific set of transcription factors is activated and these control the expression of each other as well as genes required for conidiophore morphogenesis. Recent progress has identified many of the earliest-acting genes needed for initiating conidiophore development and shown that there are at least two antagonistic signaling pathways that control this process. One pathway is modulated by a heterotrimeric G protein that when activated stimulates growth and represses both asexual and sexual sporulation as well as production of the toxic secondary metabolite, sterigmatocystin. The second pathway apparently requires an extracellular signal to induce sporulation-specific events and to direct the inactivation of the first pathway, removing developmental repression. A working model is presented in which the regulatory interactions between these two pathways during the fungal life cycle determine whether cells grow or develop.
Figures







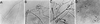
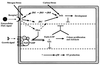
References
-
- Adams T H, Boylan M T, Timberlake W E. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell. 1988;54:353–362. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

