Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation
- PMID: 9529892
- PMCID: PMC98911
- DOI: 10.1128/MMBR.62.1.204-229.1998
Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation
Abstract
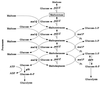
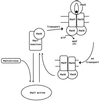
The maltose system of Escherichia coli offers an unusually rich set of enzymes, transporters, and regulators as objects of study. This system is responsible for the uptake and metabolism of glucose polymers (maltodextrins), which must be a preferred class of nutrients for E. coli in both mammalian hosts and in the environment. Because the metabolism of glucose polymers must be coordinated with both the anabolic and catabolic uses of glucose and glycogen, an intricate set of regulatory mechanisms controls the expression of mal genes, the activity of the maltose transporter, and the activities of the maltose/maltodextrin catabolic enzymes. The ease of isolating many of the mal gene products has contributed greatly to the understanding of the structures and functions of several classes of proteins. Not only was the outer membrane maltoporin, LamB, or the phage lambda receptor, the first virus receptor to be isolated, but also its three-dimensional structure, together with extensive knowledge of functional sites for ligand binding as well as for phage lambda binding, has led to a relatively complete description of this sugar-specific aqueous channel. The periplasmic maltose binding protein (MBP) has been studied with respect to its role in both maltose transport and maltose taxis. Again, the combination of structural and functional information has led to a significant understanding of how this soluble receptor participates in signaling the presence of sugar to the chemosensory apparatus as well as how it participates in sugar transport. The maltose transporter belongs to the ATP binding cassette family, and although its structure is not yet known at atomic resolution, there is some insight into the structures of several functional sites, including those that are involved in interactions with MBP and recognition of substrates and ATP. A particularly astonishing discovery is the direct participation of the transporter in transcriptional control of the mal regulon. The MalT protein activates transcription at all mal promoters. A subset also requires the cyclic AMP receptor protein for transcription. The MalT protein requires maltotriose and ATP as ligands for binding to a dodecanucleotide MalT box that appears in multiple copies upstream of all mal promoters. Recent data indicate that the ATP binding cassette transporter subunit MalK can directly inhibit MalT when the transporter is inactive due to the absence of substrate. Despite this wealth of knowledge, there are still basic issues that require clarification concerning the mechanism of MalT-mediated activation, repression by the transporter, biosynthesis and assembly of the outer membrane and inner membrane transporter proteins, and interrelationships between the mal enzymes and those of glucose and glycogen metabolism.
Figures


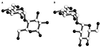
References
-
- Aiba H, Nakasa F, Mizushima S, Mizuno T. Evidence for the importance of the phosphotransfer between the two regulatory components EnvZ and OmpR in osmoregulation in Escherichia coli. J Biol Chem. 1989;264:14090–14094. - PubMed
-
- Amster-Choder O, Wright A. Transcriptional regulation of the bgl operon of Escherichia coli involves phosphotransferase system-mediated phosphorylation of a transcriptional antiterminator. J Cell Biochem. 1993;51:83–90. - PubMed
-
- Baecker P A, Furlong C E, Preiss J. Biosynthesis of bacterial glycogen. Primary structure of Escherichia coli ADP-glucose synthase as deduced from the nucleotide sequence of the glgC gene. J Biol Chem. 1983;258:5084–5088. - PubMed
-
- Baecker P A, Greenberg E, Preiss J. Biosynthesis of bacterial glycogen. Primary structure of Escherichia coli 1,4-α-d-glucan:1,4-α-d-glucan 6-α-d-(1,4-α-d-glucano)-transferase as deduced from the nucleotide sequence of the glgB gene. J Biol Chem. 1986;261:8738–8743. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

