The fission yeast SPB component Cut12 links bipolar spindle formation to mitotic control
- PMID: 9531532
- PMCID: PMC316675
- DOI: 10.1101/gad.12.7.927
The fission yeast SPB component Cut12 links bipolar spindle formation to mitotic control
Abstract
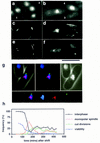
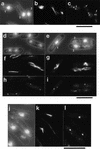
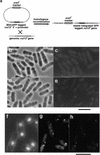
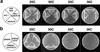
During fission yeast mitosis, the duplicated spindle pole bodies (SPBs) nucleate microtubule arrays that interdigitate to form the mitotic spindle. cut12.1 mutants form a monopolar mitotic spindle, chromosome segregation fails, and the mutant undergoes a lethal cytokinesis. The cut12(+) gene encodes a novel 62-kD protein with two predicted coiled coil regions, and one consensus phosphorylation site for p34(cdc2) and two for MAP kinase. Cut12 is localized to the SPB throughout the cell cycle, predominantly around the inner face of the interphase SPB, adjacent to the nucleus. cut12(+) is allelic to stf1(+); stf1.1 is a gain-of-function mutation bypassing the requirement for the Cdc25 tyrosine phosphatase, which normally dephosphorylates and activates the p34(cdc2)/cyclin B kinase to promote the onset of mitosis. Expressing a cut12(+) cDNA carrying the stf1.1 mutation also suppressed cdc25.22. The spindle defect in cut12.1 is exacerbated by the cdc25.22 mutation, and stf1.1 cells formed defective spindles in a cdc25.22 background at high temperatures. We propose that Cut12 may be a regulator or substrate of the p34(cdc2) mitotic kinase.
Figures




References
-
- Alfa CE, Ducommun B, Beach D, Hyams JS. Distinct nuclear and spindle pole body populations of cyclin-cdc2 in fission yeast. Nature. 1990;347:680–682. - PubMed
-
- Bailly E, Pines J, Hunter T, Bornens M. Cytoplasmic accumulation of cyclin B1 in human cells: Association with a detergent resistant compartment and with the centrosome. J Cell Sci. 1992;101:529–545. - PubMed
-
- Balczon R. The centrosome in animal cells and its functional homologs in plant and yeast cells. Int Rev Cytol. 1996;169:25–82. - PubMed
-
- Barbet N, Muriel WJ, Carr AM. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. Gene. 1992;114:59–66. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
