Translational readthrough in the hdc mRNA generates a novel branching inhibitor in the drosophila trachea
- PMID: 9531534
- PMCID: PMC316679
- DOI: 10.1101/gad.12.7.956
Translational readthrough in the hdc mRNA generates a novel branching inhibitor in the drosophila trachea
Abstract
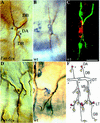
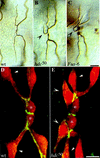
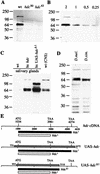
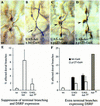
A central question in the development of many branched tubular organs, including the Drosophila trachea, concerns the mechanisms and molecules that control the number and pattern of new branches arising from preexisting vessels. We report on a branching inhibitor, Fusion-6 (Fus-6) produced by specialized tracheal cells to prevent neighboring cells from branching. In Fus-6 mutants, cells that are normally quiescent acquire the branching fate and form an increased number of sprouts emanating from the primary branches. Fus-6 is identified as the headcase (hdc) gene and is expressed in a subset of the cells that extend fusion sprouts to interconnect the tracheal network. hdc expression is regulated by the transcription factor escargot (esg) because it is not expressed in the fusion cells of esg mutants and is ectopically activated in the trachea in response to esg misexpression. We show that the hdc mRNA encodes two overlapping protein products by an unusual suppression of translational termination mechanism. Translational readthrough is necessary for hdc function because rescue of the tracheal mutant phenotype requires the full-length hdc mRNA. In ectopic expression experiments with full-length and truncated hdc constructs, only the full-length cDNA encoding both proteins could inhibit terminal branching. We propose that hdc acts non-autonomously in an inhibitory signaling mechanism to determine the number of cells that will form unicellular sprouts in the trachea.
Figures



References
-
- Ashburner M, Thompson P, Roote J, Lasko PF, Grau Y, El Messal M, Roth S, Simpson P. The genetics of a small autosomal region of Drosophila melanogaster containing the structural gene for alcohol dehydrogenase. VII. Characterization of the region around the snail and cactus loci. Genetics. 1990;126:679–694. - PMC - PubMed
-
- Böck A, Forchhammer K, Heider J, Baron C. Selenoprotein synthesis: An expansion of the genetic code. Trends Biochem Sci. 1991;16:463–467. - PubMed
-
- Bodmer R, Carretto R, Jan Y. Neurogenesis of the peripheral nervous system in Drosophila embryos: DNA replication patterns and cell lineages. Neuron. 1989;3:21–32. - PubMed
-
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
