Genes involved in osmoregulation during turgor-driven cell expansion of developing cotton fibers are differentially regulated
- PMID: 9536073
- PMCID: PMC35063
- DOI: 10.1104/pp.116.4.1539
Genes involved in osmoregulation during turgor-driven cell expansion of developing cotton fibers are differentially regulated
Abstract
Cotton (Gossypium hirsutum L.) fibers are single-celled trichomes that synchronously undergo a phase of rapid cell expansion, then a phase including secondary cell wall deposition, and finally maturation. To determine if there is coordinated regulation of gene expression during fiber expansion, we analyzed the expression of components involved in turgor regulation and a cytoskeletal protein by measuring levels of mRNA and protein accumulation and enzyme activity. Fragments of the genes for the plasma membrane proton-translocating ATPase, vacuole-ATPase, proton-translocating pyrophosphatase (PPase), phosphoenolpyruvate carboxylase, major intrinsic protein, and alpha-tubulin were amplified by polymerase chain reaction and used as probes in ribonuclease protection assays of RNA from a fiber developmental series, revealing two discrete patterns of mRNA accumulation. Transcripts of all but the PPase accumulated to highest levels during the period of peak expansion (+12-15 d postanthesis [dpa]), then declined with the onset of secondary cell wall synthesis. The PPase was constitutively expressed through fiber development. Activity of the two proton-translocating-ATPases peaked at +15 dpa, whereas PPase activity peaked at +20 dpa, suggesting that all are involved in the process of cell expansion but with varying roles. Patterns of protein accumulation and enzyme activity for some of the proteins examined suggest posttranslational regulation through fiber development.
Figures


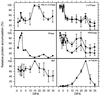
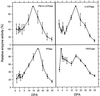
References
-
- Albert HA, Martin T, Sun SSM. Structure and expression of a sugarcane gene encoding a housekeeping phosphoenolpyruvate carboxylase. Plant Mol Biol. 1992;20:663–671. - PubMed
-
- Ames BN. Assay of inorganic phosphate, total phosphate, and phosphatases. Methods Enzymol. 1966;8:115–118.
-
- Barkla BJ, Pantoja O. Physiology of ion transport across the tonoplast of higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:159–184. - PubMed
-
- Basra AS, Malik CP. Development of the cotton fiber. Int Rev Cytol. 1984;89:65–113.
-
- Bennett AB, Leigh RA, Spanswick RM. H+-ATPase from vacuolar membranes of higher plants. Methods Enzymol. 1988;157:579–590. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

