Roles of the catalytic domain and two cellulose binding domains of Thermomonospora fusca E4 in cellulose hydrolysis
- PMID: 9537366
- PMCID: PMC107081
- DOI: 10.1128/JB.180.7.1709-1714.1998
Roles of the catalytic domain and two cellulose binding domains of Thermomonospora fusca E4 in cellulose hydrolysis
Abstract
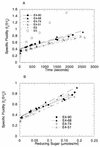
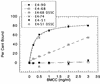
Thermomonospora fusca E4 is an unusual 90.4-kDa endocellulase comprised of a catalytic domain (CD), an internal family IIIc cellulose binding domain (CBD), a fibronectinlike domain, and a family II CBD. Constructs containing the CD alone (E4-51), the CD plus the family IIIc CBD (E4-68), and the CD plus the fibronectinlike domain plus the family II CBD (E4-74) were made by using recombinant DNA techniques. The activities of each purified protein on bacterial microcrystalline cellulose (BMCC), filter paper, swollen cellulose, and carboxymethyl cellulose were measured. Only the whole enzyme, E4-90, could reach the target digestion of 4.5% on filter paper. Removal of the internal family IIIc CBD (E4-51 and E4-74) decreased activity markedly on every substrate. E4-74 did bind to BMCC but had almost no hydrolytic activity, while E4-68 retained 32% of the activity on BMCC even though it did not bind. A low-activity mutant of one of the catalytic bases, E4-68 (Asp55Cys), did bind to BMCC, although E4-51 (Asp55Cys) did not. The ratios of soluble to insoluble reducing sugar produced after filter paper hydrolysis by E4-90, E4-68, E4-74, and E4-51 were 6.9, 3.5, 1.3, and 0.6, respectively, indicating that the family IIIc CBD is important for E4 processivity.
Figures



References
-
- Barr B K, Hsieh Y-L, Ganem B, Wilson D B. Identification of two functionally different classes of exocellulases. Biochemistry. 1996;35:586–592. - PubMed
-
- Bayer, E. A., E. Morag, R. Lamed, S. Yaron, and Y. Shoham. Cellulosome structure: four-pronged attack using biochemistry, molecular biology, crystallography and bioinformatics. In Tricel 1997 Conference Proceedings, in press. Royal Society of Chemistry, Cambridge, United Kingdom.
-
- Bothwell M, Daughhetee S, Chaua G, Wilson D B, Walker L. Binding capacities for Thermomonospora fusca E3, E4 and E5, the E3 binding domain, and Trichoderma reesei CBHI on avicel and bacterial microcrystalline cellulose. Bioresour Technol. 1997;60:169–178.
-
- Bothwell M K. Binding kinetics of Thermomonospora fusca E3 and E5, and Trichoderma reesei CBHI. Ph.D. thesis. Ithaca, N.Y: Cornell University; 1994.
-
- Chirico W J, Brown R D. Separation of [1-3H] cellooligosaccharides by thin-layer chromatography: assay for cellulolytic enzymes. Anal Biochem. 1985;150:264–272. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

