Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli
- PMID: 9537383
- PMCID: PMC107098
- DOI: 10.1128/JB.180.7.1841-1847.1998
Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli
Abstract
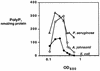
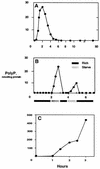
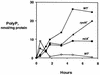
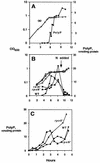
A major impediment to understanding the biological roles of inorganic polyphosphate (polyP) has been the lack of sensitive definitive methods to extract and quantitate cellular polyP. We show that polyP recovered in extracts from cells lysed with guanidinium isothiocynate can be bound to silicate glass and quantitatively measured by a two-enzyme assay: polyP is first converted to ATP by polyP kinase, and the ATP is hydrolyzed by luciferase to generate light. This nonradioactive method can detect picomolar amounts of phosphate residues in polyP per milligram of extracted protein. A simplified procedure for preparing polyP synthesized by polyP kinase is also described. Using the new assay, we found that bacteria subjected to nutritional or osmotic stress in a rich medium or to nitrogen exhaustion had large and dynamic accumulations of polyP. By contrast, carbon exhaustion, changes in pH, temperature upshifts, and oxidative stress had no effect on polyP levels. Analysis of Escherichia coli mutants revealed that polyP accumulation depends on several regulatory genes, glnD (NtrC), rpoS, relA, and phoB.
Figures

References
-
- Ahn K, Kornberg A. Polyphosphate kinase from Escherichia coli. Purification and demonstration of a phosphoenzyme intermediate. J Biol Chem. 1990;265:11734–11739. - PubMed
-
- Akiyama M, Crooke E, Kornberg A. An exopolyphosphatase of Escherichia coli. J Biol Chem. 1993;268:633–639. - PubMed
-
- Cashel M, Gentry D R, Hernandez V J, Vinella D. The stringent response. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Shaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1996. pp. 1458–1496.
-
- Chapman A G, Atkinson D E. Adenine nucleotide concentrations and turnover rates. Their correlation with biological activity in bacteria and yeast. Adv Microbiol Physiol. 1977;15:253–306. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

