The structure of the two amino-terminal domains of human ICAM-1 suggests how it functions as a rhinovirus receptor and as an LFA-1 integrin ligand
- PMID: 9539703
- PMCID: PMC22455
- DOI: 10.1073/pnas.95.8.4140
The structure of the two amino-terminal domains of human ICAM-1 suggests how it functions as a rhinovirus receptor and as an LFA-1 integrin ligand
Abstract
The normal function of human intercellular adhesion molecule-1 (ICAM-1) is to provide adhesion between endothelial cells and leukocytes after injury or stress. ICAM-1 binds to leukocyte function-associated antigen (LFA-1) or macrophage-1 antigen (Mac-1). However, ICAM-1 is also used as a receptor by the major group of human rhinoviruses and is a catalyst for the subsequent viral uncoating during cell entry. The three-dimensional atomic structure of the two amino-terminal domains (D1 and D2) of ICAM-1 has been determined to 2.2-A resolution and fitted into a cryoelectron microscopy reconstruction of a rhinovirus-ICAM-1 complex. Rhinovirus attachment is confined to the BC, CD, DE, and FG loops of the amino-terminal Ig-like domain (D1) at the end distal to the cellular membrane. The loops are considerably different in structure to those of human ICAM-2 or murine ICAM-1, which do not bind rhinoviruses. There are extensive charge interactions between ICAM-1 and human rhinoviruses, which are mostly conserved in both major and minor receptor groups of rhinoviruses. The interaction of ICAMs with LFA-1 is known to be mediated by a divalent cation bound to the insertion (I)-domain on the alpha chain of LFA-1 and the carboxyl group of a conserved glutamic acid residue on ICAMs. Domain D1 has been docked with the known structure of the I-domain. The resultant model is consistent with mutational data and provides a structural framework for the adhesion between these molecules.
Figures
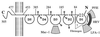



References
-
- van de Stolpe A, van der Saag P T. J Mol Med. 1996;74:13–33. - PubMed
-
- Greve J M, Davis G, Meyer A M, Forte C P, Yost S C, Marlor C W, Kamarck M E, McClelland A. Cell. 1989;56:839–847. - PubMed
-
- Staunton D E, Merluzzi V J, Rothlein R, Barton R, Marlin S D, Springer T A. Cell. 1989;56:849–853. - PubMed
-
- Berendt A R, McDowall A, Craig A G, Bates P A, Sternberg M J E, Marsh K, Newbold C I, Hogg N. Cell. 1992;68:71–81. - PubMed
-
- Ockenhouse C F, Betageri R, Springer T A, Staunton D E. Cell. 1992;68:63–69. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

